I'm trying to figure out if I can use a multilevel model for a project, and I need to simulate some data, fit a random effects model then generate the marginal effects for each group (participant or Subject in my case). However I cannot seem to generate varying marginal effects from my model fit, despite it matching a working example (e.g., the sleepstudy).
The sleepstudy provides a good starting example to work from:
library(tidyverse)
library(lme4)
library(ggeffects)
data("sleepstudy")
m <- lmer(Reaction ~ Days (1 Days | Subject), data = sleepstudy)
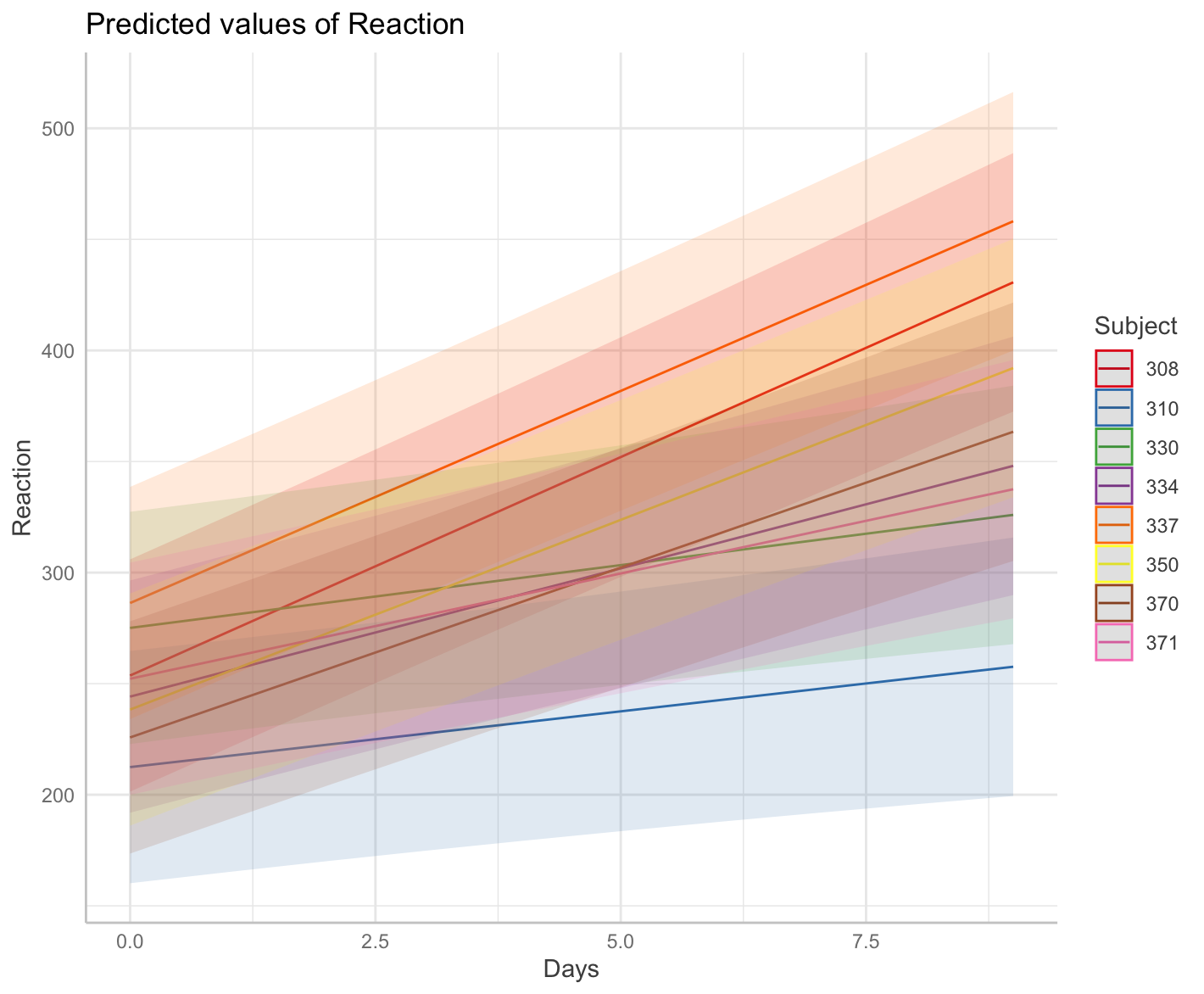
me <- ggpredict(m, terms = c("Days", "Subject [sample=8]"), type = "random")
plot(me)
The plot shows eight different slopes from eight different Subjects
However if I simulate data which approximately matches the sleep study, my model fit doesn't generate varying slopes and I don't know why not:
N = 18 # Number of people
Nt = 9 # Number of trials
d <- tibble(
Subject = factor(sprintf("d", 1:N)), # create subject numbers
subj_b0 = rnorm(n = N, mean = 250, sd = 20), # create random intercepts
subj_b1 = rnorm(n = N, mean = 10, sd = 6) # create random slopes
) %>%
mutate(Days = list(0:Nt)) %>%
unnest(Days) %>%
mutate(
Y = subj_b0 Days*subj_b1,
Y = Y rnorm(n = nrow(.), sd = 15) # Add error
)
fit <- lmer(Y ~ 1 Days (1 Days|Subject), data = d)
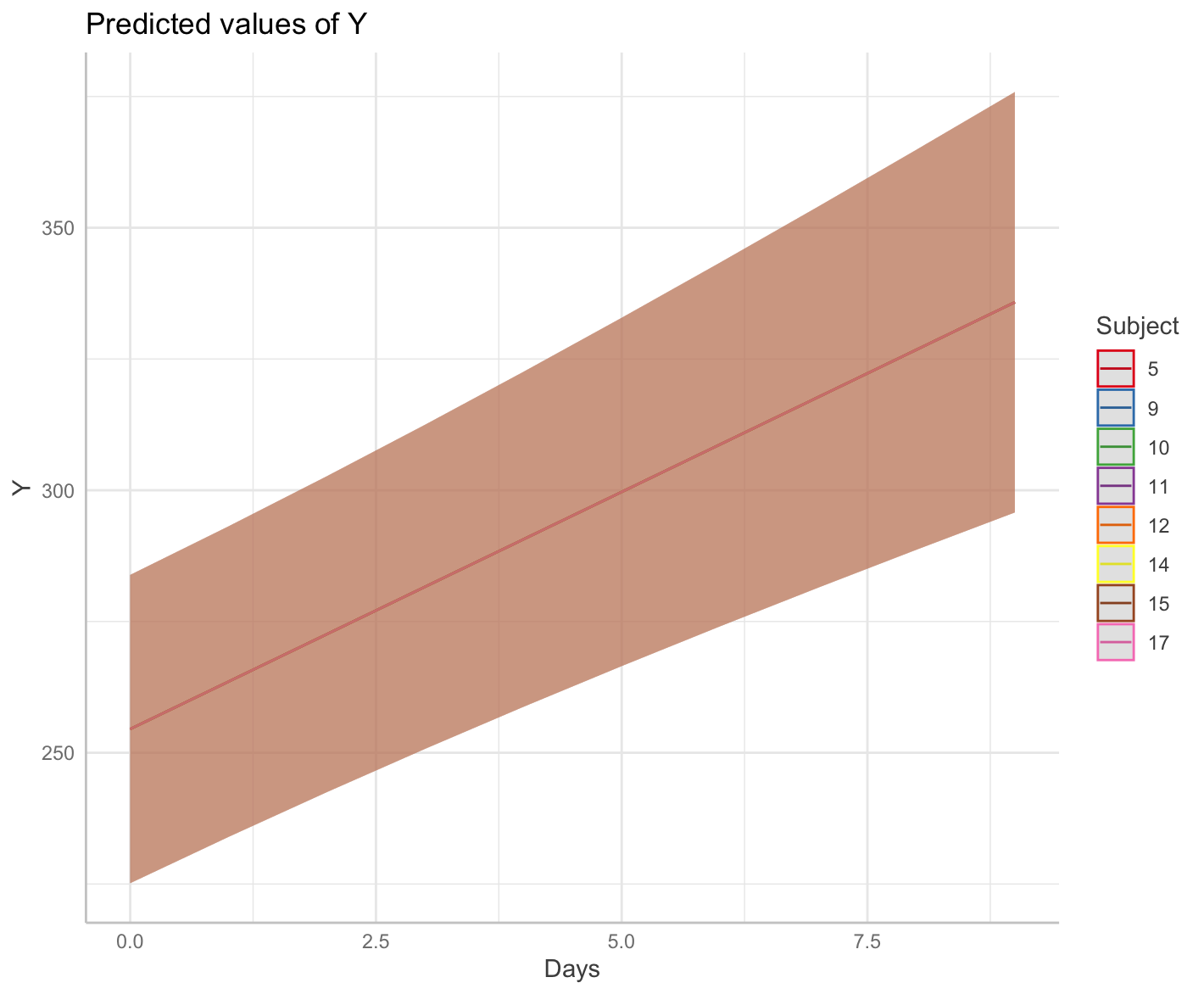
p <- ggpredict(fit, terms = c("Days", "Subject [sample=8]"), type = "random")
plot(p)
I can't for the life of me figure out why ggpredict() is returning the same slope for each Subject.
The model summary confirms there is variation around the slope for days, and as best I can tell the other estimated parameters are similar to the sleepstudy.
summary(fit)
Linear mixed model fit by REML ['lmerMod']
Formula: Y ~ 1 Days (1 Days | Subject)
Data: d
REML criterion at convergence: 1553.3
Scaled residuals:
Min 1Q Median 3Q Max
-2.57322 -0.57070 -0.01073 0.59463 3.02611
Random effects:
Groups Name Variance Std.Dev. Corr
Subject (Intercept) 745.22 27.30
Days 37.95 6.16 0.12
Residual 176.54 13.29
Number of obs: 180, groups: Subject, 18
Fixed effects:
Estimate Std. Error t value
(Intercept) 254.517 6.693 38.030
Days 9.033 1.492 6.053
sleepstudy model summary for comparison:
summary(m)
Linear mixed model fit by REML ['lmerMod']
Formula: Reaction ~ Days (1 Days | Subject)
Data: sleepstudy
REML criterion at convergence: 1743.6
Scaled residuals:
Min 1Q Median 3Q Max
-3.9536 -0.4634 0.0231 0.4634 5.1793
Random effects:
Groups Name Variance Std.Dev. Corr
Subject (Intercept) 612.10 24.741
Days 35.07 5.922 0.07
Residual 654.94 25.592
Number of obs: 180, groups: Subject, 18
Fixed effects:
Estimate Std. Error t value
(Intercept) 251.405 6.825 36.838
Days 10.467 1.546 6.771
CodePudding user response:
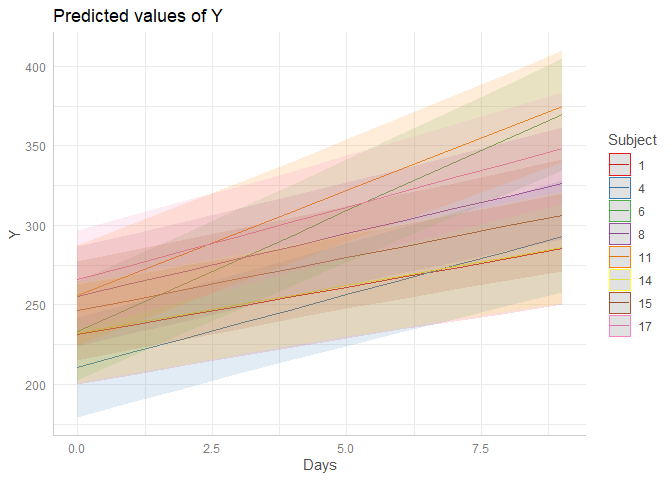
It seems that ggpredict() cannot handle factors (as random effects), which are "numeric", but have levels like 001, 002 etc. This seems to be a bug, and I will see how to fix this.
The workaround would be to change the levels for Subject:
library(ggeffects)
library(easystats)
library(lme4)
N = 18 # Number of people
Nt = 9 # Number of trials
d <- data.frame(
Subject = factor(1:N), # create subject numbers
subj_b0 = rnorm(n = N, mean = 250, sd = 20), # create random intercepts
subj_b1 = rnorm(n = N, mean = 10, sd = 6) # create random slopes
) |>
data_modify(Days = list(0:Nt)) |>
tidyr::unnest(Days) |>
data_modify(
Y = subj_b0 Days*subj_b1,
Y = Y rnorm(n = N * (Nt 1), sd = 15) # Add error
)
fit <- lmer(Y ~ 1 Days (1 Days|Subject), data = d)
p <- ggpredict(fit, terms = c("Days", "Subject [sample=8]"), type = "random")
plot(p)
Created on 2023-11-03 with reprex v2.0.2