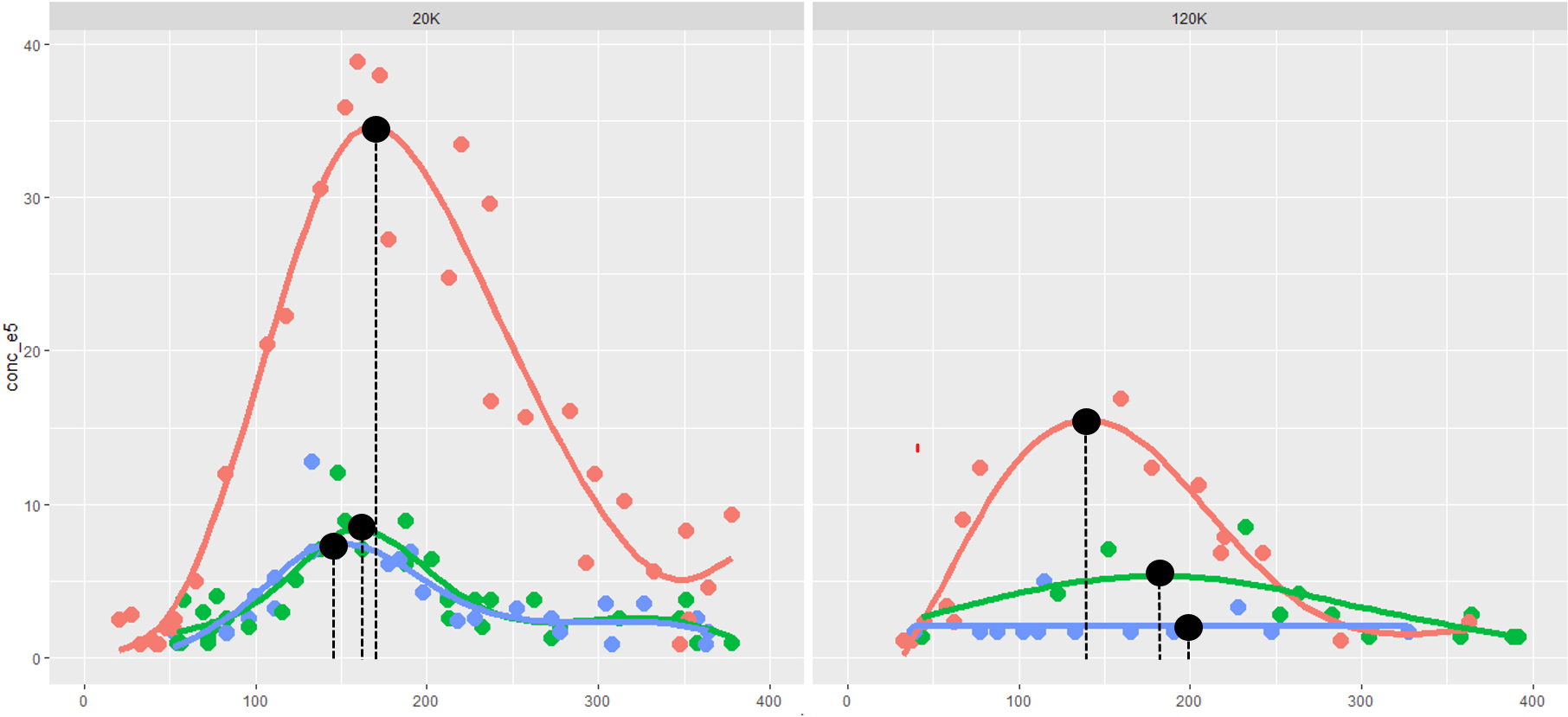
Let's say I have generated a plot with many geom_points and geom_smooth based on some strata.
I want to add a single black geom_point and black dashed line from the geom_point down to the x-axis to highlight which toppoint correspond to the x-axis value of each b$cohort within b$vesicles (used in facet_wrap).
The geom_smooth-curve should be generated with this method = "gam", formula = y ~ s(x, bs = "cs"), rather than a loess-curve.
Data
b <- structure(list(conc_e5 = c(33.5, 2, 1.1, 6.4, 1.4, 1, 1.2, 16.9,
1, 30.6, 3.3, 38, 1, 1, 7.1, 0.8, 7.1, 22.3, 1.4, 3.5, 0.9, 1,
1, 3.8, 1.2, 1.1, 0.9, 10.2, 6.2, 3.8, 1.4, 12, 1.3, 1.4, 16.7,
1.7, 5, 38.9, 2.5, 0.9, 1.7, 4.2, 2, 1.9, 2, 2.5, 5.2, 2.3, 1.4,
2.8, 4.2, 7.9, 7.1, 3.8, 1.2, 2.8, 1.7, 1.9, 1.2, 24.8, 2.6,
1.1, 3.8, 4, 6.9, 1, 4.3, 0.9, 1, 1, 6.9, 1.4, 2.8, 6.9, 4.6,
6.1, 11.3, 3.8, 3.2, 1.7, 1.1, 1.2, 0.9, 2.6, 6.8, 0.9, 3.2,
5.1, 1.4, 1.7, 1.7, 2.6, 1.3, 2.5, 8.3, 2.6, 0.9, 1.7, 2.3, 35.9,
1.7, 20.4, 2.6, 1.2, 12.8, 8.5, 5, 9, 12.4, 1, 1.2, 2.8, 1.3,
0.9, 1.7, 2.3, 1.7, 2.6, 15.7, 4, 12, 1.7, 27.3, 3, 6.8, 4, 29.6,
12.4, 3.4, 6.4, 2.8, 3.5, 9.3, 2.6, 3, 8.9, 2, 8.9, 5.6, 6.1,
12.1, 1, 2.6, 1.7, 1.7, 1.6, 1.7, 3.8, 16.1, 2.4), size = c(220,
277.5, 287.5, 202.5, 304.1, 1193, 521.7, 159.2, 722.5, 137.8,
228.1, 172.5, 762.5, 420.4, 152.5, 405.6, 162.5, 117.5, 391.2,
304.1, 432.5, 72.12, 1151, 237.5, 417.5, 37.74, 43.58, 315.3,
292.5, 58.12, 43.58, 297.5, 272.5, 407.5, 237.5, 52.17, 115.1,
159.2, 20.47, 307.5, 277.5, 263.4, 232.5, 52.17, 74.76, 52.5,
111.1, 62.46, 357.5, 283, 122.5, 220, 137.8, 227.5, 485.5, 27.5,
87.5, 47.5, 532.5, 212.5, 357.5, 557.5, 351.2, 99.7, 133, 357.5,
197.5, 472.5, 56.07, 54.08, 190.5, 387.5, 405.6, 147.5, 364.1,
187.5, 204.7, 262.5, 111.1, 39.12, 32.5, 477.5, 32.5, 347.5,
242.5, 362.5, 252.5, 123.7, 657.5, 83.29, 190.5, 212.5, 1838,
352.5, 351.2, 83.29, 347.5, 77.5, 45.18, 152.5, 247.5, 107.1,
227.5, 39.12, 132.5, 232.5, 64.74, 67.5, 177.3, 377.4, 775, 364.1,
72.12, 42.04, 165, 362.5, 111.1, 96.18, 257.5, 212.2, 82.5, 364.1,
177.5, 69.57, 217.5, 77.5, 236.4, 77.5, 57.5, 183.8, 252.5, 326.8,
377.4, 312.5, 115.1, 152.5, 96.18, 187.5, 332.5, 177.5, 148.1,
562.5, 272.5, 327.5, 132.5, 83.29, 102.5, 212.2, 283, 217.5),
cohort = c("5C", "ALL", "5C", "ALL", "ALL", "ALL", "5C",
"5C", "ALL", "5C", "KMA", "5C", "ALL", "ALL", "ALL", "KMA",
"ALL", "5C", "ALL", "KMA", "5C", "ALL", "ALL", "ALL", "5C",
"5C", "5C", "5C", "5C", "ALL", "ALL", "5C", "ALL", "ALL",
"5C", "KMA", "KMA", "5C", "5C", "KMA", "KMA", "ALL", "ALL",
"5C", "ALL", "5C", "KMA", "5C", "ALL", "ALL", "ALL", "5C",
"ALL", "ALL", "5C", "5C", "KMA", "5C", "5C", "5C", "KMA",
"5C", "ALL", "KMA", "KMA", "ALL", "KMA", "5C", "ALL", "ALL",
"KMA", "ALL", "ALL", "KMA", "5C", "ALL", "5C", "ALL", "KMA",
"KMA", "5C", "5C", "5C", "ALL", "5C", "KMA", "KMA", "ALL",
"ALL", "KMA", "KMA", "ALL", "ALL", "5C", "5C", "ALL", "5C",
"KMA", "5C", "5C", "KMA", "5C", "KMA", "5C", "KMA", "ALL",
"5C", "5C", "5C", "ALL", "5C", "ALL", "ALL", "5C", "KMA",
"5C", "KMA", "KMA", "5C", "KMA", "5C", "KMA", "5C", "ALL",
"5C", "ALL", "5C", "5C", "5C", "KMA", "ALL", "KMA", "5C",
"ALL", "ALL", "ALL", "ALL", "ALL", "5C", "KMA", "ALL", "ALL",
"KMA", "KMA", "KMA", "KMA", "KMA", "ALL", "5C", "KMA"), vesicles = structure(c(1L,
1L, 2L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, 1L,
1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L,
1L, 1L, 2L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L,
1L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L,
2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 1L, 1L,
2L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 2L,
1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, 1L, 1L, 1L, 2L,
1L, 2L, 2L, 1L, 1L, 2L, 1L, 1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 2L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, 1L, 1L), levels = c("20K",
"120K"), class = "factor")), class = "data.frame", row.names = c(NA,
-150L))
CodePudding user response:
This has been a fun learning experience for me, so is likely to be unoptimised:
# store your existing plot to an object
p <- ggplot(b, aes(y = conc_e5, x = size))
geom_point(aes(color = cohort, fill = cohort),shape = 21, size = 4, stroke = 1)
geom_smooth(method = "gam", formula = y ~ s(x, bs = "cs"),
se = FALSE, size = 2, aes(color = cohort, fill = cohort))
facet_wrap(~ vesicles)
scale_x_continuous(limits = c(0, 400))
# use ggplot2's helper function to extract the layer data
# (note - i = 2 becuase the geom_smooth is the second layer added)
pld <- layer_data(p, i = 2)
# get the maximum per panel (facet) and group (cohort)
maxes <- pld |> group_by(group, PANEL) |> slice_max(y) |> rename(cohort = group, vesicles = PANEL)
# rename the factor levels to match
levels(maxes$vesicles) <- c("20K","120K")
# plot the combined plot
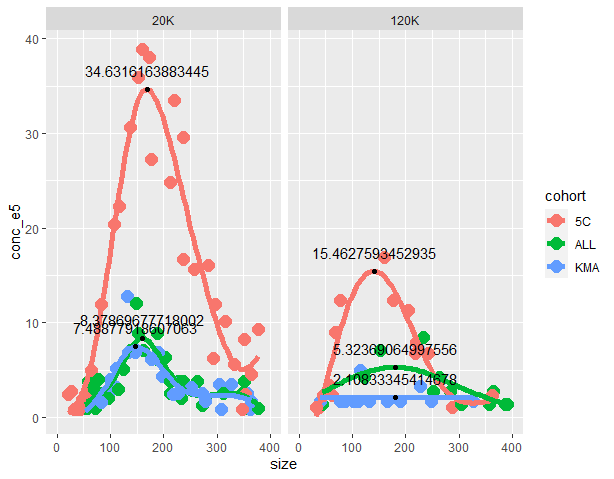
p
geom_point(data = maxes, aes(x = x, y = y))
geom_text(data = maxes, aes(x = x, y = y 2, label = x))
You can tidy up the aesthetics from there.
(note: image generated using erroneous label = y instead of x. code has been updated but image not re-generated).
CodePudding user response:
Apply the mgcv::gam() function to the data and save the maximum values for each cohort. Then add these to your existing ggplot object, like one would do when highlighting outliers:
outliers <- gapminder %>%
filter(gdpPercap >= 59000)
gapminder %>%
ggplot(aes(x = lifeExp, y = gdpPercap))
geom_point(alpha = 0.3)
geom_point(data = outliers,
aes(x = lifeExp, y = gdpPercap),
color = 'red')
Example code adapted from https://cmdlinetips.com/2019/05/how-to-highlight-select-data-points-with-ggplot2-in-r/