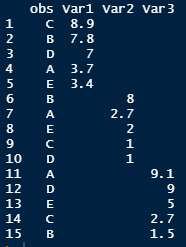
I have this dataframe:
data.frame( obs= c("A","B","C","D","E"),
Var1 = c(3.7, 7.8, 8.9, 7.0, 3.4),
Var2 = c(2.7, 8.0, 1.0, 1.0, 2.0),
Var3 = c(9.1, 1.5, 2.7, 9.0, 5.0))
to order as I need, I did it like this:
rbind(
data.frame( obs= c("A","B","C","D","E"),
Var1 = c(3.7, 7.8, 8.9, 7.0, 3.4),
Var2 = rep("",5),
Var3 = rep("",5)) %>%
arrange(-Var1),
data.frame( obs= c("A","B","C","D","E"),
Var1 = rep("",5),
Var2 = c(2.7, 8.0, 1.0, 1.0, 2.0),
Var3 = rep("",5)) %>%
arrange(-Var2),
data.frame( obs = c("A","B","C","D","E"),
Var1 = rep("",5),
Var2 = rep("",5),
Var3 = c(9.1, 1.5, 2.7, 9.0, 5.0)) %>%
arrange(-Var3)
)
output:
How to make this process more efficient and generic for multiple observations and columns?
CodePudding user response:
Get the data in long format, arrange the data in descending order, create a row number column and get the data in wide format.
Using dplyr and tidyr libraries you may do this as -
library(dplyr)
library(tidyr)
df %>%
pivot_longer(cols = -obs) %>%
arrange(name, desc(value)) %>%
mutate(row = row_number()) %>%
pivot_wider(names_from = name, values_from = value) %>%
select(-row)
# obs Var1 Var2 Var3
# <chr> <dbl> <dbl> <dbl>
# 1 C 8.9 NA NA
# 2 B 7.8 NA NA
# 3 D 7 NA NA
# 4 A 3.7 NA NA
# 5 E 3.4 NA NA
# 6 B NA 8 NA
# 7 A NA 2.7 NA
# 8 E NA 2 NA
# 9 C NA 1 NA
#10 D NA 1 NA
#11 A NA NA 9.1
#12 D NA NA 9
#13 E NA NA 5
#14 C NA NA 2.7
#15 B NA NA 1.5
CodePudding user response:
A fast solution using Map and just the base package.
r <- do.call(rbind,
Map(\(x, y, z) {x <- x[y, ]; x[-c(1, z)] <- NA; x},
list(d), lapply(d[-1], order, decreasing=T), seq(ncol(d))[-1]))
r
# obs Var1 Var2 Var3
# 3 C 8.9 NA NA
# 2 B 7.8 NA NA
# 4 D 7.0 NA NA
# 1 A 3.7 NA NA
# 5 E 3.4 NA NA
# 21 B NA 8.0 NA
# 11 A NA 2.7 NA
# 51 E NA 2.0 NA
# 31 C NA 1.0 NA
# 41 D NA 1.0 NA
# 12 A NA NA 9.1
# 42 D NA NA 9.0
# 52 E NA NA 5.0
# 32 C NA NA 2.7
# 22 B NA NA 1.5
Benchmark
This should be slightly more efficient.
# Unit: microseconds
# expr min lq mean median uq max neval cld
# Map 876.396 900.545 970.8851 981.9155 1014.284 1145.8 100 a
# dplyr 14744.610 14963.718 17997.4444 15439.0440 15752.575 241073.9 100 b
Data:
d <- structure(list(obs = c("A", "B", "C", "D", "E"), Var1 = c(3.7,
7.8, 8.9, 7, 3.4), Var2 = c(2.7, 8, 1, 1, 2), Var3 = c(9.1, 1.5,
2.7, 9, 5)), class = "data.frame", row.names = c(NA, -5L))