hy, Here is an update proposal for the solution presented by 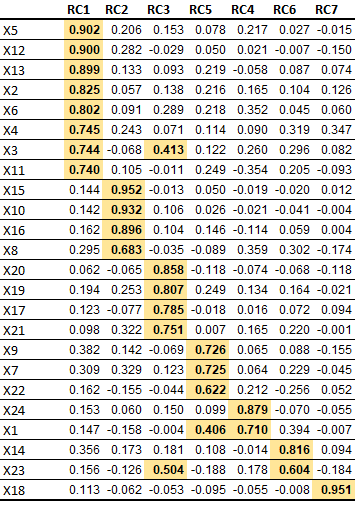
CodePudding user response:
Now the question is cleared up, I think this does what you want:
library(dplyr)
df %>%
mutate(across(everything(),
~ ifelse(. < 0.4, "", format(., digits = 3)))) %>%
arrange(across(everything(), desc))
# RC1 RC2 RC3 RC4 RC5 RC6 RC7
# 1 0.902
# 2 0.900
# 3 0.899
# 4 0.825
# 5 0.802
# 6 0.745
# 7 0.744 0.413
# 8 0.740
# 9 0.952
# 10 0.932
# 11 0.896
# 12 0.683
# 13 0.858
# 14 0.807
# 15 0.785
# 16 0.751
# 17 0.504 0.604
# 18 0.726
# 19 0.725
# 20 0.622
# 21 0.406 0.710
# 22 0.878
# 23 0.816
# 24 0.951
CodePudding user response:
library(tidyverse)
data <-
data.frame(
id=c("X5","X12","X13","X2","X6", "X4","X3","X11","X15","X10","X16","X8","X20","X19","X17","X21","X9","X7","X22","X24","X1","X14","X23","X18"),
RC1=c(0.902,0.9,0.899,0.825,0.802,0.745,0.744,0.74,0.382,0.356,0.309,0.295,0.194,0.162,0.162,0.156,0.153,0.147,0.144,0.142,0.123,0.113,0.098,0.062),
RC2=c(0.206,0.282,0.133,0.057,0.091,0.243,-0.068,0.105,0.143,0.173,0.329,0.683,0.253,0.896,-0.155,-0.126,0.06,-0.158,0.952,0.932,-0.077,-0.062,0.322,-0.065),
RC3=c(0.153,-0.029,0.093,0.138,0.289,0.071,0.413,-0.011,-0.069,0.181,0.123,-0.035,0.807,0.104,-0.044,0.504,0.15,-0.004,-0.013,0.106,0.785,-0.053,0.751,0.858),
RC4=c(0.078,0.05,0.219,0.216,0.218,0.114,0.122,0.249,0.726,0.108,0.725,-0.089,0.249,0.146,0.622,-0.189,0.099,0.406,0.05,0.026,-0.018,-0.095,0.007,-0.118),
RC5=c(0.217,0.021,-0.058,0.166,0.352,0.09,0.26,-0.354,0.065,-0.014,0.064,0.359,0.134,-0.114,0.212,0.178,0.878,0.71,-0.019,-0.021,0.015,-0.055,0.165,-0.074),
RC6=c(0.027,-0.007,0.087,0.104,0.045,0.319,0.296,0.205,0.088,0.816,0.229,0.302,0.163,0.059,-0.256,0.604,-0.07,0.394,-0.02,-0.041,0.071,-0.008,0.219,-0.068),
RC7=c(-0.015,-0.15,0.073,0.126,0.06,0.347,0.082,-0.093,-0.155,0.093,-0.045,-0.175,-0.021,0.004,0.052,-0.184,-0.054,-0.008,0.012,-0.004,0.094,0.951,-0.001,-0.118)
)
# Question 1: How to sort the columns, from largest to smallest, in each column, as in the image?
data %>% arrange(-RC1)
#> id RC1 RC2 RC3 RC4 RC5 RC6 RC7
#> 1 X5 0.902 0.206 0.153 0.078 0.217 0.027 -0.015
#> 2 X12 0.900 0.282 -0.029 0.050 0.021 -0.007 -0.150
#> 3 X13 0.899 0.133 0.093 0.219 -0.058 0.087 0.073
#> 4 X2 0.825 0.057 0.138 0.216 0.166 0.104 0.126
#> 5 X6 0.802 0.091 0.289 0.218 0.352 0.045 0.060
#> 6 X4 0.745 0.243 0.071 0.114 0.090 0.319 0.347
#> 7 X3 0.744 -0.068 0.413 0.122 0.260 0.296 0.082
#> 8 X11 0.740 0.105 -0.011 0.249 -0.354 0.205 -0.093
#> 9 X15 0.382 0.143 -0.069 0.726 0.065 0.088 -0.155
#> 10 X10 0.356 0.173 0.181 0.108 -0.014 0.816 0.093
#> 11 X16 0.309 0.329 0.123 0.725 0.064 0.229 -0.045
#> 12 X8 0.295 0.683 -0.035 -0.089 0.359 0.302 -0.175
#> 13 X20 0.194 0.253 0.807 0.249 0.134 0.163 -0.021
#> 14 X19 0.162 0.896 0.104 0.146 -0.114 0.059 0.004
#> 15 X17 0.162 -0.155 -0.044 0.622 0.212 -0.256 0.052
#> 16 X21 0.156 -0.126 0.504 -0.189 0.178 0.604 -0.184
#> 17 X9 0.153 0.060 0.150 0.099 0.878 -0.070 -0.054
#> 18 X7 0.147 -0.158 -0.004 0.406 0.710 0.394 -0.008
#> 19 X22 0.144 0.952 -0.013 0.050 -0.019 -0.020 0.012
#> 20 X24 0.142 0.932 0.106 0.026 -0.021 -0.041 -0.004
#> 21 X1 0.123 -0.077 0.785 -0.018 0.015 0.071 0.094
#> 22 X14 0.113 -0.062 -0.053 -0.095 -0.055 -0.008 0.951
#> 23 X23 0.098 0.322 0.751 0.007 0.165 0.219 -0.001
#> 24 X18 0.062 -0.065 0.858 -0.118 -0.074 -0.068 -0.118
# Question 2: How to hide the values in each column when the value is =< 0.04?
data %>% filter(RC1 > 0.04)
#> id RC1 RC2 RC3 RC4 RC5 RC6 RC7
#> 1 X5 0.902 0.206 0.153 0.078 0.217 0.027 -0.015
#> 2 X12 0.900 0.282 -0.029 0.050 0.021 -0.007 -0.150
#> 3 X13 0.899 0.133 0.093 0.219 -0.058 0.087 0.073
#> 4 X2 0.825 0.057 0.138 0.216 0.166 0.104 0.126
#> 5 X6 0.802 0.091 0.289 0.218 0.352 0.045 0.060
#> 6 X4 0.745 0.243 0.071 0.114 0.090 0.319 0.347
#> 7 X3 0.744 -0.068 0.413 0.122 0.260 0.296 0.082
#> 8 X11 0.740 0.105 -0.011 0.249 -0.354 0.205 -0.093
#> 9 X15 0.382 0.143 -0.069 0.726 0.065 0.088 -0.155
#> 10 X10 0.356 0.173 0.181 0.108 -0.014 0.816 0.093
#> 11 X16 0.309 0.329 0.123 0.725 0.064 0.229 -0.045
#> 12 X8 0.295 0.683 -0.035 -0.089 0.359 0.302 -0.175
#> 13 X20 0.194 0.253 0.807 0.249 0.134 0.163 -0.021
#> 14 X19 0.162 0.896 0.104 0.146 -0.114 0.059 0.004
#> 15 X17 0.162 -0.155 -0.044 0.622 0.212 -0.256 0.052
#> 16 X21 0.156 -0.126 0.504 -0.189 0.178 0.604 -0.184
#> 17 X9 0.153 0.060 0.150 0.099 0.878 -0.070 -0.054
#> 18 X7 0.147 -0.158 -0.004 0.406 0.710 0.394 -0.008
#> 19 X22 0.144 0.952 -0.013 0.050 -0.019 -0.020 0.012
#> 20 X24 0.142 0.932 0.106 0.026 -0.021 -0.041 -0.004
#> 21 X1 0.123 -0.077 0.785 -0.018 0.015 0.071 0.094
#> 22 X14 0.113 -0.062 -0.053 -0.095 -0.055 -0.008 0.951
#> 23 X23 0.098 0.322 0.751 0.007 0.165 0.219 -0.001
#> 24 X18 0.062 -0.065 0.858 -0.118 -0.074 -0.068 -0.118
# Question 3:That the solution is, if possible, generic for n columns
data %>% filter_at(vars(starts_with("RC")), ~ .x > 0.04)
#> id RC1 RC2 RC3 RC4 RC5 RC6 RC7
#> 1 X2 0.825 0.057 0.138 0.216 0.166 0.104 0.126
#> 2 X6 0.802 0.091 0.289 0.218 0.352 0.045 0.060
#> 3 X4 0.745 0.243 0.071 0.114 0.090 0.319 0.347
# Question 4: If possible, how visually can the doR output be presented in table format (expected output)?.
# Output is already a table, you can use kable package for HTML table rendering
Created on 2021-09-09 by the reprex package (v2.0.1)
