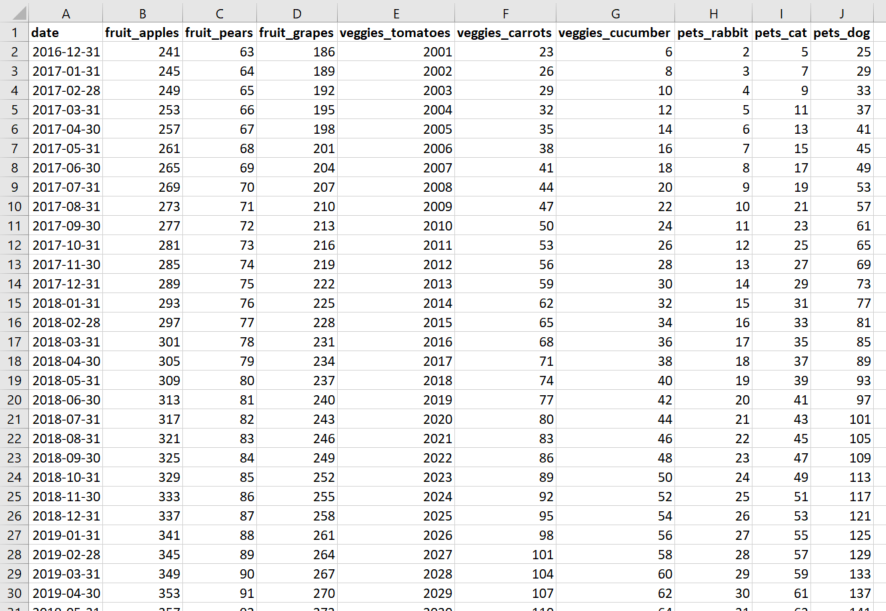
I have a huge times series dataset (over 500 variables) consisting of monthly observations for several years. See a simplified example of the input data below (in Excel):
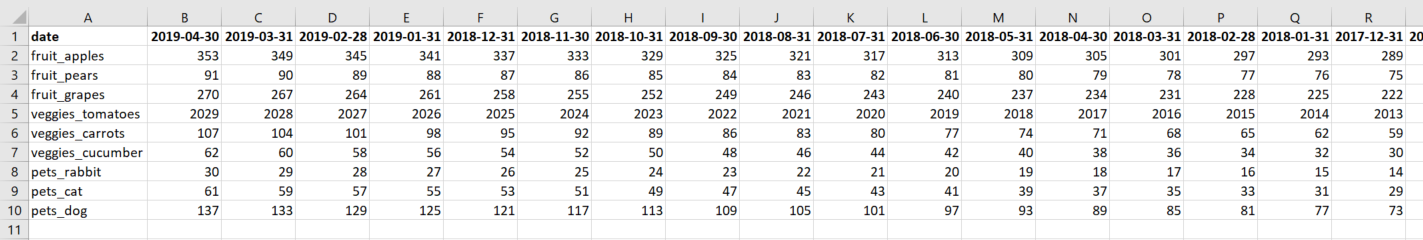
Now, what I need to do (in R) is to transpose this input data set so that the variable date ends up as the new variable names and the current variable names end up in the column date. See a simplified example of the desired output data below (in Excel):
Note that is important that the dates in the variable names in the desired end result need to be in descending order (newest closest to the left).
I do manage to transpose the data using the function t(), however, the problem comes after that.
First problem: The variable date is automatically set as rownames after transposing, which is fine, I just run the function rownames_to_column() to fix this. However, is there a way to avoid this maneuver?
Second problem: All the new variable names end up in the first row, but when I run the function row_to_names(1) to change this the dates in the variable names end up weird. The only solution to this that I have made work (almost all the way) is to first convert all variables to character format before running row_to_names(1). After that I need to change my date-variables back to numeric format which I need to do in order to proceed with further calculations. However, when I do this on my real data set (which is the same just much bigger) I get the error message: "NAs introduced by coercion". Why does this happen? I can't seem to find out why. If you find a different approach to do this that is also fine.
# Example data to copy and paste into R for easy reproduction of problem:
df <- data.frame (date = c("2018-11-30", "2018-12-31", "2019-01-31", "2019-02-28", "2019-03-31", "2019-04-30"),
fruit_apples = c(333, 337, 341, 345, 349, 353),
fruit_pears = c(86, 87, 88, 89, 90, 91),
fruit_grapes = c(255, 258, 261, 264, 267, 270),
veggies_tomatoes = c(2024, 2025, 2026, 2027, 2028, 2029),
veggies_carrots = c(92, 95, 98, 101, 104, 107),
veggies_cucumber = c(52, 54, 56, 58, 60, 62),
pets_rabbit = c(25, 26, 27, 28, 29, 30),
pets_cat = c(51, 53, 55, 57, 59, 61),
pets_dog = c(117, 121, 125, 129, 133, 137))
# The code I have been using so far:
df_output <- df %>%
arrange(desc(date)) %>% # after transposing I want the newest date to be closest to the left, hence this step
t() %>%
as.data.frame() %>%
rownames_to_column() %>%
mutate(across(everything(), as.character)) %>%
row_to_names(1) %>%
mutate_at(vars(-date), as.numeric) %>% # causes error message "NAs introduced by coercion"
adorn_totals()
CodePudding user response:
I hope this is what you have in mind:
library(tidyr)
df %>%
pivot_longer(!date, names_to = "Date", values_to = "value") %>%
pivot_wider(names_from = date, values_from = value)
# A tibble: 9 x 7
Date `2018-11-30` `2018-12-31` `2019-01-31` `2019-02-28` `2019-03-31` `2019-04-30`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 fruit_apples 333 337 341 345 349 353
2 fruit_pears 86 87 88 89 90 91
3 fruit_grapes 255 258 261 264 267 270
4 veggies_tomatoes 2024 2025 2026 2027 2028 2029
5 veggies_carrots 92 95 98 101 104 107
6 veggies_cucumber 52 54 56 58 60 62
7 pets_rabbit 25 26 27 28 29 30
8 pets_cat 51 53 55 57 59 61
9 pets_dog 117 121 125 129 133 137
CodePudding user response:
A data.table way -
library(data.table)
dcast(melt(setDT(df), id.vars = 'date'), variable~date, value.var = 'value')
# variable 2018-11-30 2018-12-31 2019-01-31 2019-02-28 2019-03-31 2019-04-30
#1: fruit_apples 333 337 341 345 349 353
#2: fruit_pears 86 87 88 89 90 91
#3: fruit_grapes 255 258 261 264 267 270
#4: veggies_tomatoes 2024 2025 2026 2027 2028 2029
#5: veggies_carrots 92 95 98 101 104 107
#6: veggies_cucumber 52 54 56 58 60 62
#7: pets_rabbit 25 26 27 28 29 30
#8: pets_cat 51 53 55 57 59 61
#9: pets_dog 117 121 125 129 133 137