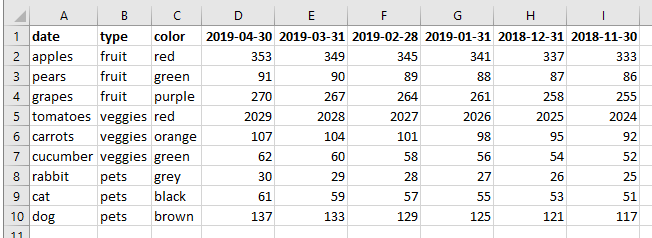
I have a dataset in R where some of the variable names are dates, see a simplified example of the input data below (in Excel):
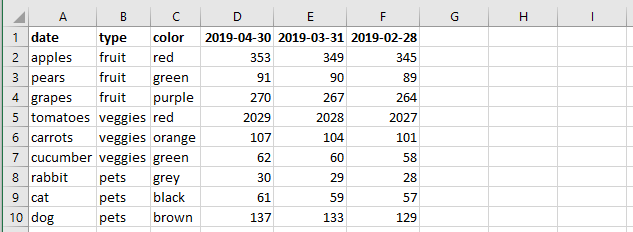
What I want to do with this data is to remove some of the columns with names that are dates that are older than or equal to a certain date, e.g. 2019-01-31. See a simplified example of the desired output data below (in Excel):
Now, I am able to achieve this by transposing the data, filtering out rows with a date lower than or equal to 31 January 2019 and finally transposing the data back. However I am wondering whether there is a different way to do this using just the column names without pivoting back and forth?
# Example data to copy and paste into R for easy reproduction of problem:
df <- data.frame (id = c("apples", "pears", "grapes", "tomatoes", "carrots", "cucumber", "rabbit", "cat", "dog"),
type = c("fruit", "fruit", "fruit", "veggies", "veggies", "veggies", "pets", "pets", "pets"),
color = c("red", "green", "purple", "red", "orange", "green", "grey", "black", "brown"),
'2019-04-30' = c(353, 91, 270, 2029, 107, 62, 30, 61, 137),
'2019-03-31' = c(349, 90, 267, 2028, 104, 60, 29, 59, 133),
'2019-02-28' = c(345, 89, 264, 2027, 101, 58, 28, 57, 129),
'2019-01-31' = c(341, 88, 261, 2026, 98, 56, 27, 55, 125),
'2018-12-31' = c(337, 87, 258, 2025, 95, 54, 26, 53, 121),
'2018-11-30' = c(333, 86, 255, 2024, 92, 52, 25, 51, 117),
check.names = FALSE)
CodePudding user response:
The approach is as follows:
- extract the column names
- transform to
Dateif possible andNAif not date like - create boolean vector to filter too old dates and non dates (i.e.
NAsin the step before) columns
Sample Data
## sample data frame
m <- matrix(1, 3, 10)
colnames(m) <- c("a", "b", as.character(seq.Date(as.Date("2021-1-1"), length.out = 8, by = "days")))
(d <- as.data.frame(m))
# a b 2021-01-01 2021-01-02 2021-01-03 2021-01-04 2021-01-05 2021-01-06 2021-01-07 2021-01-08
# 1 1 1 1 1 1 1 1 1 1 1
# 2 1 1 1 1 1 1 1 1 1 1
# 3 1 1 1 1 1 1 1 1 1 1
Filter
r <- vapply(names(d), as.Date, numeric(1), optional = TRUE)
d[, is.na(r) | r <= as.Date("2021-1-3")]
# a b 2021-01-01 2021-01-02 2021-01-03
# 1 1 1 1 1 1
# 2 1 1 1 1 1
# 3 1 1 1 1 1
r <- vapply(names(df), as.Date, numeric(1), optional = TRUE)
df[, is.na(r) | r >= as.Date("2019-1-31")]
# id type color 2019-04-30 2019-03-31 2019-02-28 2019-01-31
# 1 apples fruit red 353 349 345 341
# 2 pears fruit green 91 90 89 88
# 3 grapes fruit purple 270 267 264 261
# 4 tomatoes veggies red 2029 2028 2027 2026
# 5 carrots veggies orange 107 104 101 98
# 6 cucumber veggies green 62 60 58 56
# 7 rabbit pets grey 30 29 28 27
# 8 cat pets black 61 59 57 55
# 9 dog pets brown 137 133 129 125
CodePudding user response:
We can do this in base R. Your dates are conveniently in YYYY-MM-DD format, which means they will be ordered correctly by the >= and <= operators. We can also use a simple regex to preserve any columns that are not in date format:
df[!grepl('\\d{4}-\\d{2}-\\d{2}', colnames(df)) | colnames(df) >= '2019-02-28']
id type color 2019-04-30 2019-03-31 2019-02-28
1 apples fruit red 353 349 345
2 pears fruit green 91 90 89
3 grapes fruit purple 270 267 264
4 tomatoes veggies red 2029 2028 2027
5 carrots veggies orange 107 104 101
6 cucumber veggies green 62 60 58
7 rabbit pets grey 30 29 28
8 cat pets black 61 59 57
9 dog pets brown 137 133 129
CodePudding user response:
Description
One can re-shape the data to the long format and filter based on the date column.
Data
Same data as provided in the example
df <- data.frame (id = c("apples", "pears", "grapes", "tomatoes", "carrots", "cucumber", "rabbit", "cat", "dog"),
type = c("fruit", "fruit", "fruit", "veggies", "veggies", "veggies", "pets", "pets", "pets"),
color = c("red", "green", "purple", "red", "orange", "green", "grey", "black", "brown"),
'2019-04-30' = c(353, 91, 270, 2029, 107, 62, 30, 61, 137),
'2019-03-31' = c(349, 90, 267, 2028, 104, 60, 29, 59, 133),
'2019-02-28' = c(345, 89, 264, 2027, 101, 58, 28, 57, 129),
'2019-01-31' = c(341, 88, 261, 2026, 98, 56, 27, 55, 125),
'2018-12-31' = c(337, 87, 258, 2025, 95, 54, 26, 53, 121),
'2018-11-30' = c(333, 86, 255, 2024, 92, 52, 25, 51, 117),
check.names = FALSE)
Solution
library(dplyr)
library(tidyr)
df %>%
tidyr::pivot_longer(cols = !c(id, type, color), names_to = 'date', values_to = 'value') %>%
dplyr::mutate(date = as.Date(date, format = '%Y-%m-%d')) %>%
dplyr::filter( date >= as.Date('2019-01-31')) %>%
tidyr::pivot_wider(names_from = 'date', values_from = 'value')
Desired output
id type color `2019-04-30` `2019-03-31` `2019-02-28` `2019-01-31`
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 apples fruit red 353 349 345 341
2 pears fruit green 91 90 89 88
3 grapes fruit purple 270 267 264 261
4 tomatoes veggies red 2029 2028 2027 2026
5 carrots veggies orange 107 104 101 98
6 cucumber veggies green 62 60 58 56
7 rabbit pets grey 30 29 28 27
8 cat pets black 61 59 57 55
9 dog pets brown 137 133 129 125