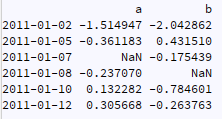
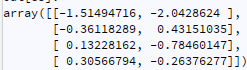
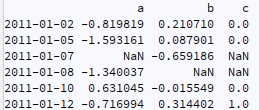
I have a time series like following:
from datetime import datetime
dates = [datetime(2011, 1, 2), datetime(2011, 1, 5), datetime(2011, 1, 7), datetime(2011, 1, 8), datetime(2011, 1, 10), datetime(2011, 1, 12)]
ts = pd.DataFrame({"a":np.random.randn(6),"b":np.random.randn(6)}, index=dates)
ts.iloc[2,0]=np.nan
ts.iloc[3,1]=np.nan
so it happens on many instances that we need to convert it to numpy array, with nonull values, and do different processis, like NN, etc...
ts.dropna().values
Now for example lets say that a new column c is generated from numpy array calculations(clustering, NN,...):
what is the best way to add this to original df so it becomes like:
in other words in this workflow:
1- start with pandas dataframe multifeature time series
2- remove nulls
3- calculate a new array from 2 (classification, NN, ...)
4- adding the array created in 3 to the original dataframe in step 1 (how to do this properly?)
I know that some might say we can stick with pandas for the entire process, but lets say the table gets 3 dimensional that we need to convert it to numpy array.
Thank you!
CodePudding user response:
Try isna/notna to mask your data, then .loc to assign back:
valids = ts.notna().all(axis=1)
# equivalent to ts.dropna().values
data = ts[valids].to_numpy()
# do stuff
preds = KMeans().fit_predict(data)
# preds = [0, 0, 0, 1]
# assign prediction back
# ravel in the case your predictions are 2D as shown
ts.loc[valids, 'pred'] = preds.ravel()
CodePudding user response:
- drop your NaNs from the dataframe and assign the index to a variable.
- create a pandas dataframe containing
c, with this index - left join this new dataframe to the original