So I am tasked with building an excel spreadsheet cataloging a drive with various nested folders and files.
This 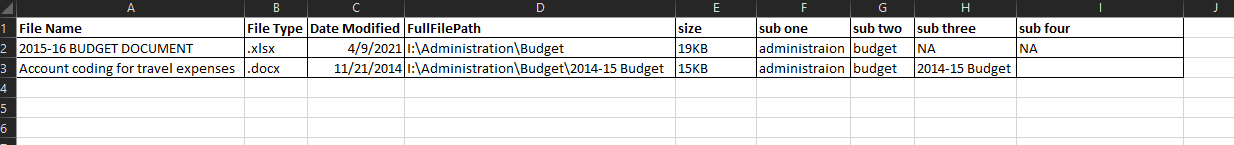
I know that there might be a command to get file info and I can break that into these columns.
CodePudding user response:
Apart from the directories split into subdirs, the adaptation of the function in the question's link, Stibu's answer, might be of help.
rfl <- function(path) {
folders <- list.dirs(path, recursive = FALSE, full.names = FALSE)
if (length(folders)==0) {
files <- list.files(path, full.names = TRUE)
finfo <- file.info(files)
Filename <- basename(files)
FileType <- tools::file_ext(files)
DateModified <- finfo$mtime
FullFilePath <- dirname(files)
size <- finfo$size
data.frame(Filename, FileType, DateModified, FullFilePath, size)
} else {
sublist <- lapply(paste0(path,"/",folders),rfl)
setNames(sublist,folders)
}
}
CodePudding user response:
If you have the full path and file names then you can loop through that and parse it into these columns. You can get more file info with file.info:
files <- c("I:/Administration/Budget/2015-BUDGET DOCUMENT.xlsx",
"I:/Administration/Budget/2014-2015 Budget/BUDGET DOCUMENT.xlsx")
# files <- list.files("I:", recursive = T, full.names = T) # this could take a while to run
file_info <- list(length = length(files))
for (i in seq_along(files)){
fullpath <- dirname(files[i])
fullname <- basename(files[i])
file_ext <- unlist(strsplit(fullname, ".", fixed = T))
file_meta <- file.info(files[i])[c("size", "mtime")]
path <- unlist(strsplit(fullpath, "/", fixed = T))[-1]
file_info[[i]] <- unlist(c(file_ext, file_meta, fullpath, path))
}
l <- lapply(file_info, `length<-`, max(lengths(file_info)))
df <- data.frame(do.call(rbind, l))
names(df) <- c("filename", "extension", "size", "modified", paste0("sub", 1:(ncol(df) - 4)))
rownames(df) <- NULL
df$modified <- as.POSIXct.numeric(as.numeric(df$modified), origin = "1970-01-01")
df$size <- as.numeric(df$size)
If you do not have the files you can recursively search the drive using list.files() with recursive = T: list.files("I:", recursive = T, full.names = T)
Note:
l <- lapply(file_info, `length<-`, max(lengths(file_info)))sets the vector length of each list element to be the same. This is necessary because otherwise when the vectors are stacked with unequal lengths values get recycled. A simple example of this is:rbind(1:3, 1:5)- The output of
unlist(c(file_ext, file_meta, fullpath, path))is a vector and vectors in R are atomic, meaning all elements have to be the same class. That means everything gets converted to character in this case, which is why we have the linesdf$modified <- ...anddf$size <- ...at the end to convert them to their appropriate type. - If you want to output this data frame to excel check out
xlsx::write.xlsxoropenxlsx::write.xlsx. If you don't have those libraries installed you'll need to useinstall.packages()first.
Output
Because these files/locations don't actually exist on my computer there are NA values in the size and date modified fields:
filename extension size modified sub1 sub2 sub3 sub4
1 2015-BUDGET DOCUMENT xlsx NA <NA> I:/Administration/Budget Administration Budget <NA>
2 BUDGET DOCUMENT xlsx NA <NA> I:/Administration/Budget/2014-2015 Budget Administration Budget 2014-2015 Budget
