I'm trying to do a rowwise matching for vector elements nested inside a data.table list-column.
library(tibble)
library(data.table)
my_dt <-
tibble::tribble(
~col_x, ~col_y,
"a", c(1, 2, 3),
"b", c(4, 5, 6),
"c", c(7, 8, 9)
) %>%
as.data.table()
external_vec <- 1:9
I want to match each element from col_y against external_vec such that I will get a new column in my_dt that is equivalent to external_vec %in% col_y, such as:
# desired output
## col_x col_y new_col
## 1: a 1,2,3 TRUE, TRUE, TRUE,FALSE,FALSE,FALSE,...
## 2: b 4,5,6 FALSE,FALSE,FALSE, TRUE, TRUE, TRUE,...
## 3: c 7,8,9 FALSE,FALSE,FALSE,FALSE,FALSE,FALSE,...
EDIT
I know how to implement a solution based on tidyverse's purrr, like the following. However, I'm looking for a data.table native functionality.
# this works but relies on purrr and dplyr, which I hope to avoid in this problem
my_dt %>%
tibble::add_column(ext_vec_as_col = list(external_vec)) %>%
mutate(new_col = purrr::map2(.x = ext_vec_as_col, .y = col_y, .f = ~.x %in% .y) )
#> col_x col_y ext_vec_as_col new_col
#> 1: a 1,2,3 1,2,3,4,5,6,... TRUE, TRUE, TRUE,FALSE,FALSE,FALSE,...
#> 2: b 4,5,6 1,2,3,4,5,6,... FALSE,FALSE,FALSE, TRUE, TRUE, TRUE,...
#> 3: c 7,8,9 1,2,3,4,5,6,... FALSE,FALSE,FALSE,FALSE,FALSE,FALSE,...
CodePudding user response:
I've got the tidyverse-solution:
library(tibble)
library(dplyr)
library(tidyr)
library(purrr)
library(magrittr)
my_dt <-
tibble::tribble(
~col_x, ~col_y,
"a", c(1, 2, 3),
"b", c(4, 5, 6),
"c", c(7, 8, 9)
)
external_vec <- 1:9
my_dt %>%
mutate(new_col = col_y %>%
map(~external_vec %in% .x)) %>%
unnest_wider(col_y, names_sep = "_") %>% # this is just to see the result..
unnest_wider(new_col, names_sep = "_") # this is just to see the result..
Usually, library(tidyverse) includes a lot of these, but I just thought that it would be nicer to know exactly what you need to make this work.
data.table solution
I think this is the equivalent:
my_dt[, new_col := lapply(col_y, \(x) external_vec %in% x)]
my_dt
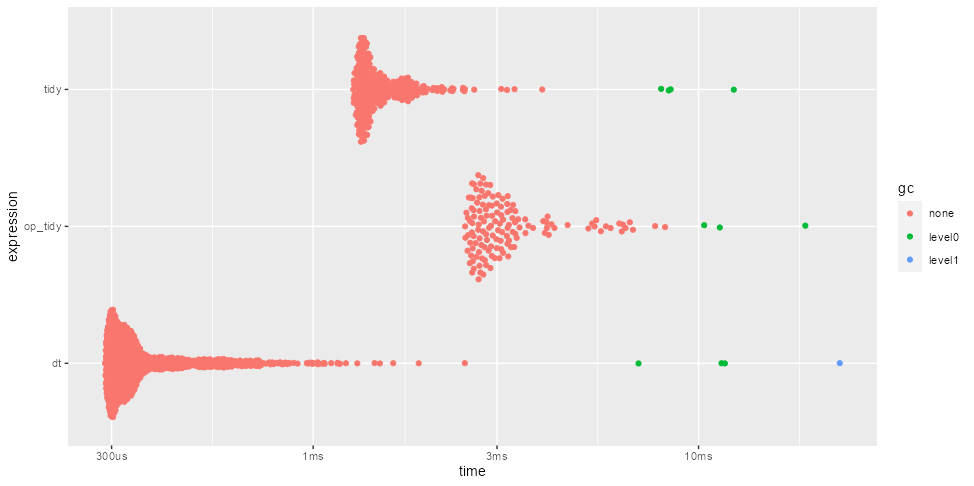
Performance
Since that is important, here is some performance benchmark
bench::mark(
tidy =
my_dt %>%
mutate(new_col = col_y %>%
map(~external_vec %in% .x)),
dt = my_dt[, new_col := lapply(col_y, \(x) external_vec %in% x)],
op_tidy = my_dt %>%
tibble::add_column(ext_vec_as_col = list(external_vec)) %>%
mutate(new_col = purrr::map2(.x = ext_vec_as_col, .y = col_y, .f = ~.x %in% .y)),
check = FALSE
) %>%
plot()