I'm wondering if there's a way to quickly calculate the number of shared neighbors between any pairs of nodes (i.e., the number of nodes connected to both node i and j) from an adjacency matrix (like the one below) and then return the output in matrix format?
I've referenced these following posts,
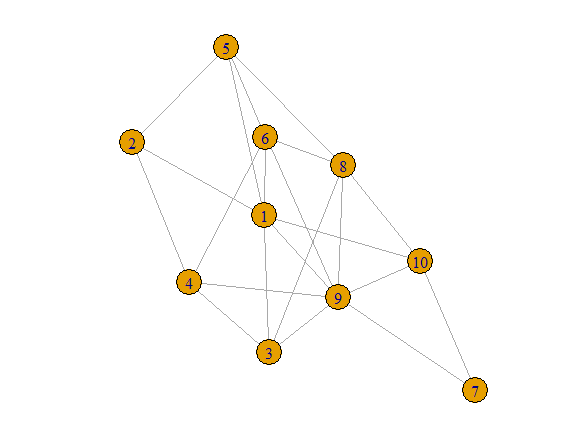
Notice for example that Shared[1,4] = 4.
That is because nodes 1 and 4 have four shared neighbors,
Nodes 2,3,6 and 9. Shared[5,7]=0 because nodes 5 and 7
have no neighbors in common.
CodePudding user response:
I think @G5W's solution is super concise already, highly recommended!
Below is a graph theory way to solve it, maybe not that efficient:
> nb <- neighborhood(g,mindist = 1)
> outer(nb, nb, FUN = Vectorize(function(a, b) length(intersect(a, b))))
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
[1,] 6 1 1 4 2 2 2 5 3 1
[2,] 1 3 2 0 1 3 0 1 2 1
[3,] 1 2 4 1 2 4 1 1 3 3
[4,] 4 0 1 4 2 1 1 3 2 1
[5,] 2 1 2 2 4 2 0 1 3 2
[6,] 2 3 4 1 2 5 1 2 3 3
[7,] 2 0 1 1 0 1 2 2 1 1
[8,] 5 1 1 3 1 2 2 5 3 1
[9,] 3 2 3 2 3 3 1 3 7 3
[10,] 1 1 3 1 2 3 1 1 3 4
> t(adjm) %*% adjm
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
[1,] 6 1 1 4 2 2 2 5 3 1
[2,] 1 3 2 0 1 3 0 1 2 1
[3,] 1 2 4 1 2 4 1 1 3 3
[4,] 4 0 1 4 2 1 1 3 2 1
[5,] 2 1 2 2 4 2 0 1 3 2
[6,] 2 3 4 1 2 5 1 2 3 3
[7,] 2 0 1 1 0 1 2 2 1 1
[8,] 5 1 1 3 1 2 2 5 3 1
[9,] 3 2 3 2 3 3 1 3 7 3
[10,] 1 1 3 1 2 3 1 1 3 4
