I've got a time series of sunspot numbers, where the mean number of sunspots is counted per month, and I'm trying to use a Fourier Transform to convert from the time domain to the frequency domain. The data used is from https://wwwbis.sidc.be/silso/infosnmtot. The first thing I'm confused about is how to express the sampling frequency as once per month. Do I need to convert it to seconds, eg. 1/(seconds in 30 days)? Here's what I've got so far:
fs = 1/2592000
#the sampling frequency is 1/(seconds in a month)
fourier = np.fft.fft(sn_value)
#sn_value is the mean number of sunspots measured each month
freqs = np.fft.fftfreq(sn_value.size,d=fs)
power_spectrum = np.abs(fourier)
plt.plot(freqs,power_spectrum)
plt.xlim(0,max(freqs))
plt.title("Power Spectral Density of the Sunspot Number Time Series")
plt.grid(True)
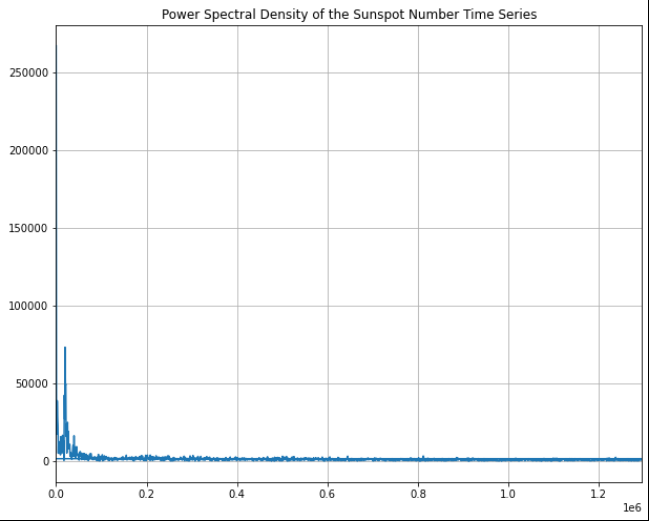
I don't think this is correct - namely because I don't know what the scale of the x-axis is. However I do know that there should be a peak at (11years)^-1.
The second thing I'm wondering from this graph is why there seems to be two lines - one being a horizontal line just above y=0. It's more clear when I change the x-axis bounds to: plt.xlim(0,1).
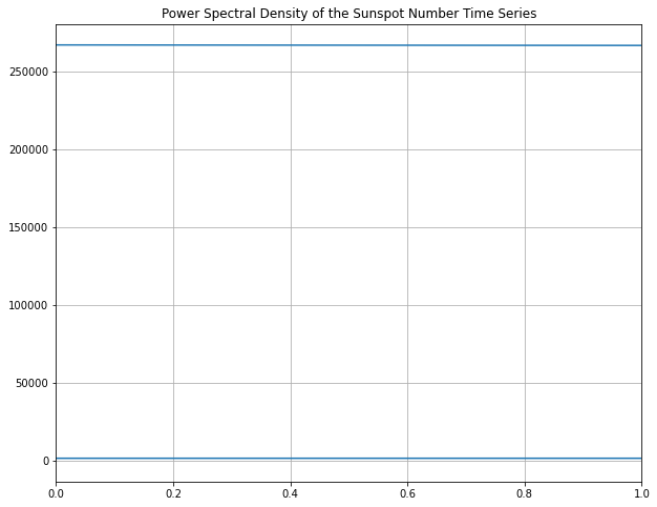
Am I using the fourier transform functions incorrectly?
CodePudding user response:
You can use any units you want. Feel free to express your sampling frequency in samples/year. The x-axis will then have 1/year units.
The extra line you spotted comes from the way you plot your data. Look at the output of the np.fft.fftfreq call. The first half of that array contains positive values from 0 to 1.2e6 or so, the other half contain negative values from -1.2e6 to almost 0. By plotting all your data, you get a data line from 0 to the right, then a straight line from the rightmost point to the leftmost point, then the rest of the data line back to zero. Your xlim call makes it so you don’t see half the data plotted.
Typically you’d plot only the first half of your data, just crop the freqs and power_spectrum arrays.
