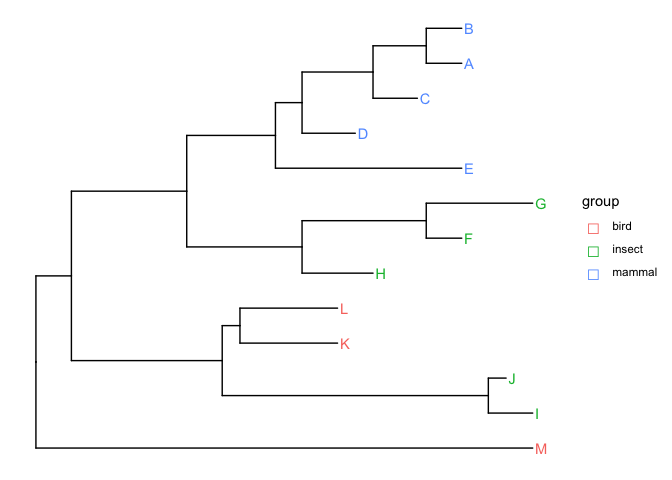
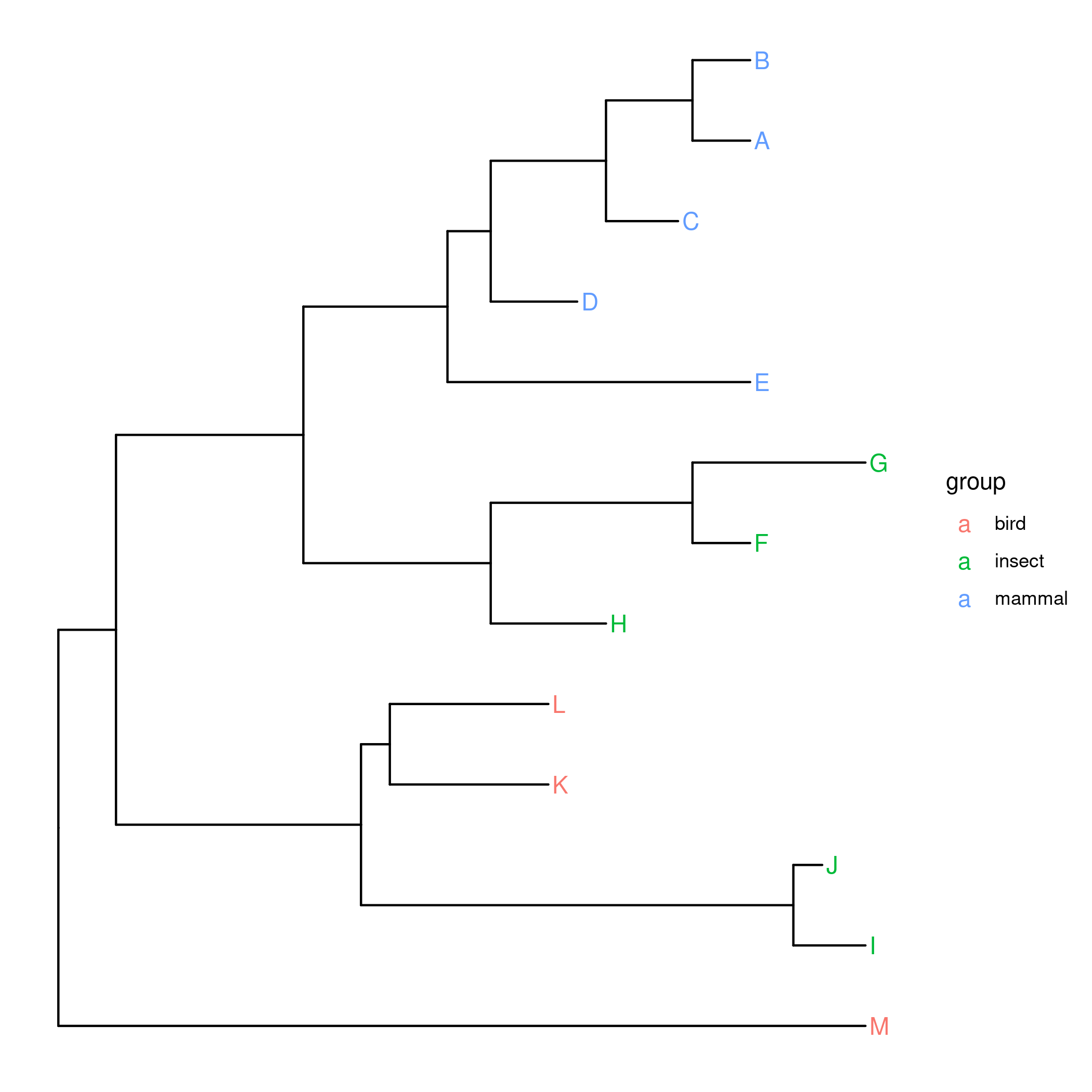
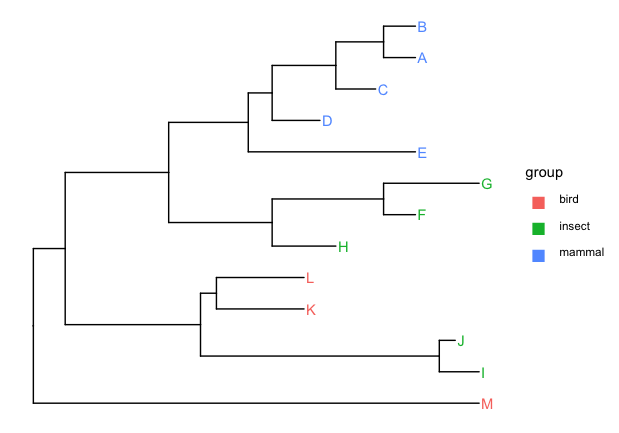
Given a tree with tip labels colored by a group eg:
library(ggplot2)
library(ggtree)
nwk <- system.file("extdata", "sample.nwk", package="treeio")
tree <- read.tree(nwk)
meta = data.frame(label = c("A", "B", "C", "D", "E", "F", "G", "H", "I", "J", "K", "L", "M"),
group = c(rep("mammal", 5), rep("insect", 5), rep("bird", 3)))
p = ggtree(tree) %< % meta
geom_tiplab(aes(color = group))
The legend will contain a instead of the desired square. ggplot2 documentation says to use "override.aes" to override this behavior:
p guides(color = guide_legend(override.aes = list(size = 4,
label = "",
shape = 15)))
This does not work. The overriding works, I can change the size, colors, remove the a but most importantly I can't use a different shape (in this case a square).
Newer version of ggplot2 have the key_glyph option, but this too has no effect when using geom_tiplab.
Another odd behavior is that when using geom_tipppoint, overriding the legend works. Of note, is that the above strategies previously worked to remove the a, however in the latest R/ggplot2/ggtree the above mentioned strategies do not work. This is related to the latest version of ggtree and not older versions.
Any advice on how to override geom_tiplab() legend shape?
My environment:
R version 4.1.2 (2021-11-01)
ggplot2 version 3.3.5
ggtree version 3.2.1
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Linux Mint 20
CodePudding user response:
I'm not sure if there's a clever way to do this, but a potential workaround is to change the label to the unicode 'square' character, e.g.
library(tidyverse)
library(ggtree)
#> ggtree v3.0.4 For help: https://yulab-smu.top/treedata-book/
#>
#> If you use ggtree in published research, please cite the most appropriate paper(s):
#>
#> 1. Guangchuang Yu. Using ggtree to visualize data on tree-like structures. Current Protocols in Bioinformatics, 2020, 69:e96. doi:10.1002/cpbi.96
#> 2. Guangchuang Yu, Tommy Tsan-Yuk Lam, Huachen Zhu, Yi Guan. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Molecular Biology and Evolution 2018, 35(12):3041-3043. doi:10.1093/molbev/msy194
#> 3. Guangchuang Yu, David Smith, Huachen Zhu, Yi Guan, Tommy Tsan-Yuk Lam. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 2017, 8(1):28-36. doi:10.1111/2041-210X.12628
#>
#> Attaching package: 'ggtree'
#> The following object is masked from 'package:tidyr':
#>
#> expand
nwk <- system.file("extdata", "sample.nwk", package="treeio")
tree <- read.tree(nwk)
meta = data.frame(label = c("A", "B", "C", "D", "E", "F", "G", "H", "I", "J", "K", "L", "M"),
group = c(rep("mammal", 5), rep("insect", 5), rep("bird", 3)))
p = ggtree(tree) %< % meta
geom_tiplab(aes(color = group))
guides(color = guide_legend(override.aes = list(label = "\u25A1", size = 6)))
p