I have a dataset containing time periods during which an intervention is happening. We have two types of interventions. I have the start and end date of each intervention. I would now like to extract the time (in days) when there is no overlap between the two types and how much overlap there is.
Here's an example dataset:
data <- data.table( id = seq(1,21),
type = as.character(c(1,2,2,2,2,2,2,2,1,1,1,1,1,2,1,2,1,1,1,1,1)),
start_dt = as.Date(c("2015-01-09", "2015-04-14", "2015-06-19", "2015-10-30", "2016-03-01", "2016-05-24",
"2016-08-03", "2017-08-18", "2017-08-18", "2018-02-01", "2018-05-07", "2018-08-09",
"2019-01-31", "2019-03-22", "2019-05-16", "2019-11-04", "2019-11-04", "2020-02-06",
"2020-05-28", "2020-08-25", "2020-12-14")),
end_dt = as.Date(c("2017-07-24", "2015-05-04", "2015-08-27", "2015-11-19", "2016-03-21", "2016-06-09",
"2017-07-18", "2019-02-21", "2018-01-23", "2018-04-25", "2018-07-29", "2019-01-15",
"2019-04-24", "2019-09-13", "2019-10-13", "2020-12-23", "2020-01-26", "2020-04-29",
"2020-08-19", "2020-11-16", "2021-03-07")))
> data
id type start_dt end_dt
1: 1 1 2015-01-09 2017-07-24
2: 2 2 2015-04-14 2015-05-04
3: 3 2 2015-06-19 2015-08-27
4: 4 2 2015-10-30 2015-11-19
5: 5 2 2016-03-01 2016-03-21
6: 6 2 2016-05-24 2016-06-09
7: 7 2 2016-08-03 2017-07-18
8: 8 2 2017-08-18 2019-02-21
9: 9 1 2017-08-18 2018-01-23
10: 10 1 2018-02-01 2018-04-25
11: 11 1 2018-05-07 2018-07-29
12: 12 1 2018-08-09 2019-01-15
13: 13 1 2019-01-31 2019-04-24
14: 14 2 2019-03-22 2019-09-13
15: 15 1 2019-05-16 2019-10-13
16: 16 2 2019-11-04 2020-12-23
17: 17 1 2019-11-04 2020-01-26
18: 18 1 2020-02-06 2020-04-29
19: 19 1 2020-05-28 2020-08-19
20: 20 1 2020-08-25 2020-11-16
21: 21 1 2020-12-14 2021-03-07
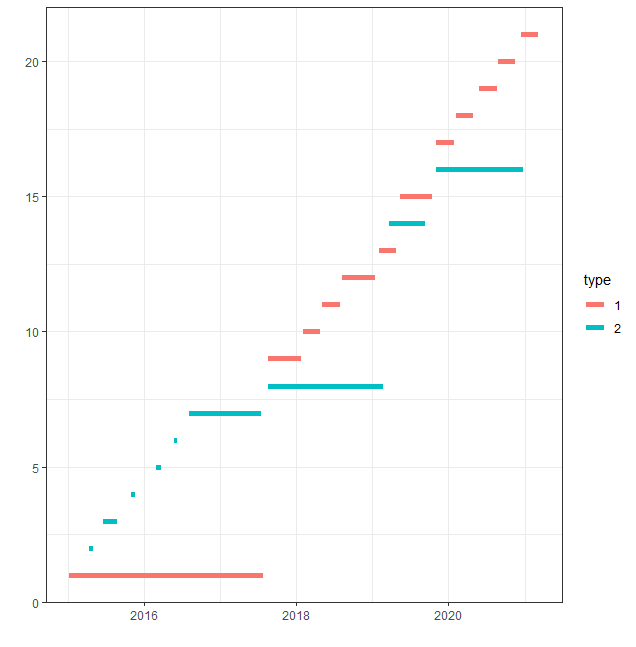
Here's a plot of the data for a better view of what I want to know:
library(ggplot2)
ggplot(data = data,
aes(x = start_dt, xend = end_dt, y = id, yend = id, color = type))
geom_segment(size = 2)
xlab("")
ylab("")
theme_bw()
I'll describe the first part of the example: we have an intervention of type 1 from 2015-01-09 until 2017-07-24. From 2015-04-14 however, also intervention type 2 is happening. This means that we only have "pure" type 1 from 2015-01-09 to 2015-04-13, which is 32 days.
Then we have an overlapping period from 2015-04-14 to 2015-05-04, which is 20 days. Then we again have a period with only type 1 from 2015-05-05 to 2015-06-18, which is 44 days. In total, we now have had (32 44 =) 76 days of "pure" type 1 and 20 days of overlap. Then we continue like this for the entire time period.
I would like to know the total time (in days) of "pure" type 1, "pure" type 2 and overlap.
Alternatively, if also possible, I would like to organise the data such, that I get all the seperate time periods extracted, meaning that the data would look something like this (type 3 = overlap):
> data_adjusted
id type start_dt end_dt
1: 1 1 2015-01-09 2015-04-14
2: 2 3 2015-04-15 2015-05-04
3: 3 1 2015-05-05 2015-06-18
4: 4 3 2015-06-19 2015-08-27
........
The time in days spent in each intervention type can then easily be calculated from data_adjuted.
I have similar answers using dplyr or just marking overlapping time periods, but I have not found an answer to my specific case.
Is there an efficient way to calculate this using data.table?
CodePudding user response:
This method does a small explosion of looking at all dates in the range, so it may not scale very well if your data gets large.
library(data.table)
alldates <- data.table(date = seq(min(data$start_dt), max(data$end_dt), by = "day"))
data[alldates, on = .(start_dt <= date, end_dt >= date)] %>%
.[, .N, by = .(start_dt, type) ] %>%
.[ !is.na(type), ] %>%
dcast(start_dt ~ type, value.var = "N") %>%
.[, r := do.call(rleid, .SD), .SDcols = setdiff(colnames(.), "start_dt") ] %>%
.[, .(type = fcase(is.na(`1`[1]), "2", is.na(`2`[1]), "1", TRUE, "3"),
start_dt = min(start_dt), end_dt = max(start_dt)), by = r ]
# r type start_dt end_dt
# <int> <char> <Date> <Date>
# 1: 1 1 2015-01-09 2015-04-13
# 2: 2 3 2015-04-14 2015-05-04
# 3: 3 1 2015-05-05 2015-06-18
# 4: 4 3 2015-06-19 2015-08-27
# 5: 5 1 2015-08-28 2015-10-29
# 6: 6 3 2015-10-30 2015-11-19
# 7: 7 1 2015-11-20 2016-02-29
# 8: 8 3 2016-03-01 2016-03-21
# 9: 9 1 2016-03-22 2016-05-23
# 10: 10 3 2016-05-24 2016-06-09
# 11: 11 1 2016-06-10 2016-08-02
# 12: 12 3 2016-08-03 2017-07-18
# 13: 13 1 2017-07-19 2017-07-24
# 14: 14 3 2017-08-18 2018-01-23
# 15: 15 2 2018-01-24 2018-01-31
# 16: 16 3 2018-02-01 2018-04-25
# 17: 17 2 2018-04-26 2018-05-06
# 18: 18 3 2018-05-07 2018-07-29
# 19: 19 2 2018-07-30 2018-08-08
# 20: 20 3 2018-08-09 2019-01-15
# 21: 21 2 2019-01-16 2019-01-30
# 22: 22 3 2019-01-31 2019-02-21
# 23: 23 1 2019-02-22 2019-03-21
# 24: 24 3 2019-03-22 2019-04-24
# 25: 25 2 2019-04-25 2019-05-15
# 26: 26 3 2019-05-16 2019-09-13
# 27: 27 1 2019-09-14 2019-10-13
# 28: 28 3 2019-11-04 2020-01-26
# 29: 29 2 2020-01-27 2020-02-05
# 30: 30 3 2020-02-06 2020-04-29
# 31: 31 2 2020-04-30 2020-05-27
# 32: 32 3 2020-05-28 2020-08-19
# 33: 33 2 2020-08-20 2020-08-24
# 34: 34 3 2020-08-25 2020-11-16
# 35: 35 2 2020-11-17 2020-12-13
# 36: 36 3 2020-12-14 2020-12-23
# 37: 37 1 2020-12-24 2021-03-07
# r type start_dt end_dt
It drops the id field, I don't know how to map it well back to your original data.
CodePudding user response:
@r2evans solution is more complete, but if you want to explore the use offoverlaps you can start with something like this:
#split into two frames
data = split(data,by="type")
# key the second frame
setkey(data[[2]], start_dt, end_dt)
# create the rows that have overlaps
overlap = foverlaps(data[[1]],data[[2]], type="any", nomatch=0)
# get the overlapping time periods
overlap[, .(start_dt = max(start_dt,i.start_dt), end_dt=min(end_dt,i.end_dt)), by=1:nrow(overlap)][,type:=3]
Output:
nrow start_dt end_dt type
1: 1 2015-04-14 2015-05-04 3
2: 2 2015-06-19 2015-08-27 3
3: 3 2015-10-30 2015-11-19 3
4: 4 2016-03-01 2016-03-21 3
5: 5 2016-05-24 2016-06-09 3
6: 6 2016-08-03 2017-07-18 3
7: 7 2017-08-18 2018-01-23 3
8: 8 2018-02-01 2018-04-25 3
9: 9 2018-05-07 2018-07-29 3
10: 10 2018-08-09 2019-01-15 3
11: 11 2019-01-31 2019-02-21 3
12: 12 2019-03-22 2019-04-24 3
13: 13 2019-05-16 2019-09-13 3
14: 14 2019-11-04 2020-01-26 3
15: 15 2020-02-06 2020-04-29 3
16: 16 2020-05-28 2020-08-19 3
17: 17 2020-08-25 2020-11-16 3
18: 18 2020-12-14 2020-12-23 3
The sum of those overlap days is 1492.