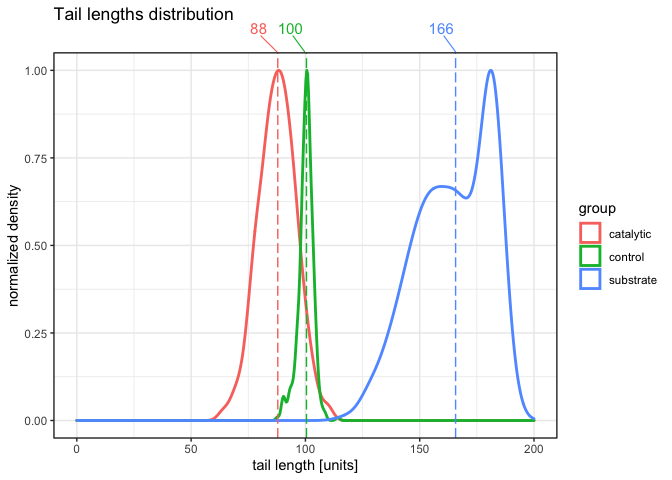
In would like to plot density plots with certain values (for instance: median/mean/etc.). I also would like to display chosen values (for instance median) above the plotting area, so it would not interfere with the distributions itself. Also, in real life I have larger, more diverse dataframes (with much more categories) so I would like to spread the labels, so they would not interfere with each other (I want them to be readable and visually pleasing).
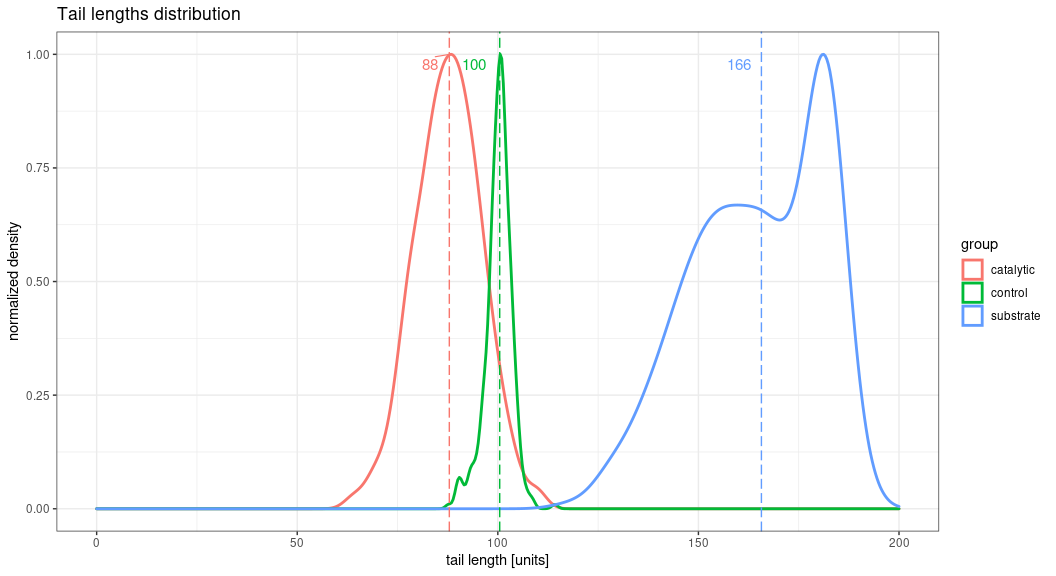
And this is the output I would love to have (sorry for the quality, edited in paint):
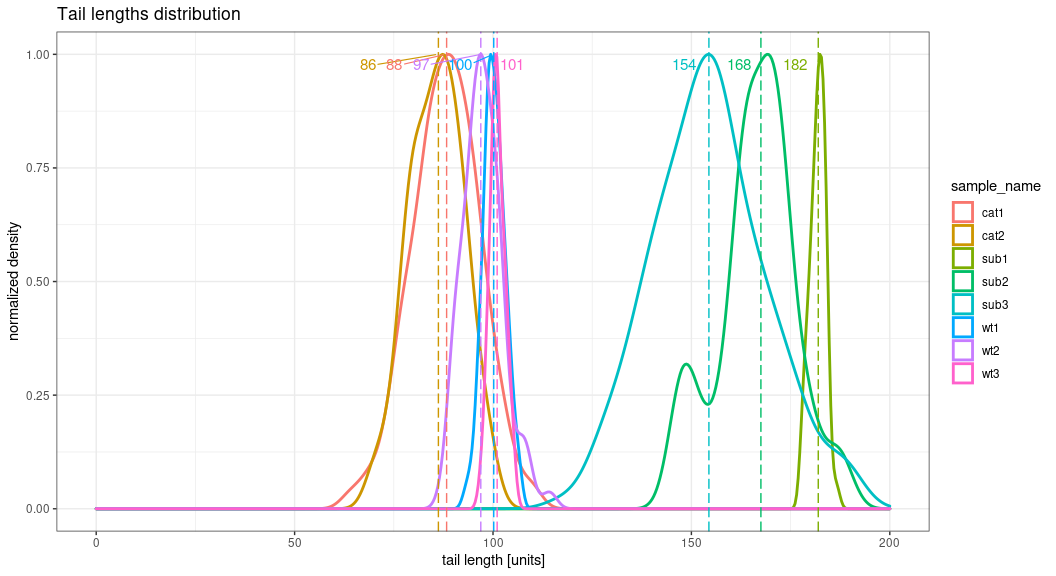
Also, if you change the grouping factor for "sample_name", then you will see more "crowded" plot, more similar to my irl data.
CodePudding user response:
One option to achieve your desired result:
- Set
clip="off" incoord_cartesian` - Make some room for the labels by increasing the bottom margin of the title
- Set
y=1.05for the labels (the max of data range the default expansion of the scale by .05) - Set
min.segment.length=0 - Increase the
ylimfor the labels - Nudge the position of the labels
Note: Getting your desired result you probably have to fiddle around with the values for the nudging, the ylim and the margin.
set.seed(123)
library(ggplot2)
library(ggrepel)
library(dplyr)
plot_distribution_with_values <- function(input_data,value_to_show="mean", grouping_factor = "group", title="", limit="") {
#determine the center values to be plotted as x intercepting line(s)
center_values = input_data %>% dplyr::group_by(!!rlang::sym(grouping_factor)) %>% dplyr::summarize(median_value = median(tail_length,na.rm = TRUE),mean_value=mean(tail_length,na.rm=T))
#main core of the plot
plot_distribution <- ggplot2::ggplot(input_data, aes_string(x=tail_length,color=grouping_factor))
geom_density(size=1, aes(y=..ndensity..)) theme_bw() scale_x_continuous(limits=c(0, as.numeric(limit)))
coord_cartesian(clip = "off", ylim = c(0, 1))
if (value_to_show=="median") {
center_value="median_value"
}
else if (value_to_show=="mean") {
center_value="mean_value"
}
#Plot settings (aesthetics, geoms, axes behavior etc.):
g.line <- ggplot2::geom_vline(data=center_values,aes(xintercept=!!rlang::sym(center_value),
color=!!rlang::sym(grouping_factor)),
linetype="longdash",show.legend = FALSE)
g.labs <- ggplot2::labs(title= "Tail lengths distribution",
x="tail length [units]",
y= "normalized density",
color=grouping_factor)
g.values <- ggrepel::geom_text_repel(data=center_values,
aes(x=round(!!rlang::sym(center_value)),
y = 1.05, color=!!rlang::sym(grouping_factor),
label=formatC(round(!!rlang::sym(center_value)),digits=1,format = "d")),
size=4, direction = "x", segment.size = 0.4,
min.segment.length = 0, nudge_y = .15, nudge_x = -10,
show.legend =F, hjust =0, xlim = c(0,200),
ylim = c(0, 1.15))
#Overall plotting configuration:
plot <- plot_distribution g.line g.labs g.values
theme(plot.title = element_text(margin = margin(b = 4 * 5.5)))
return(plot)
}
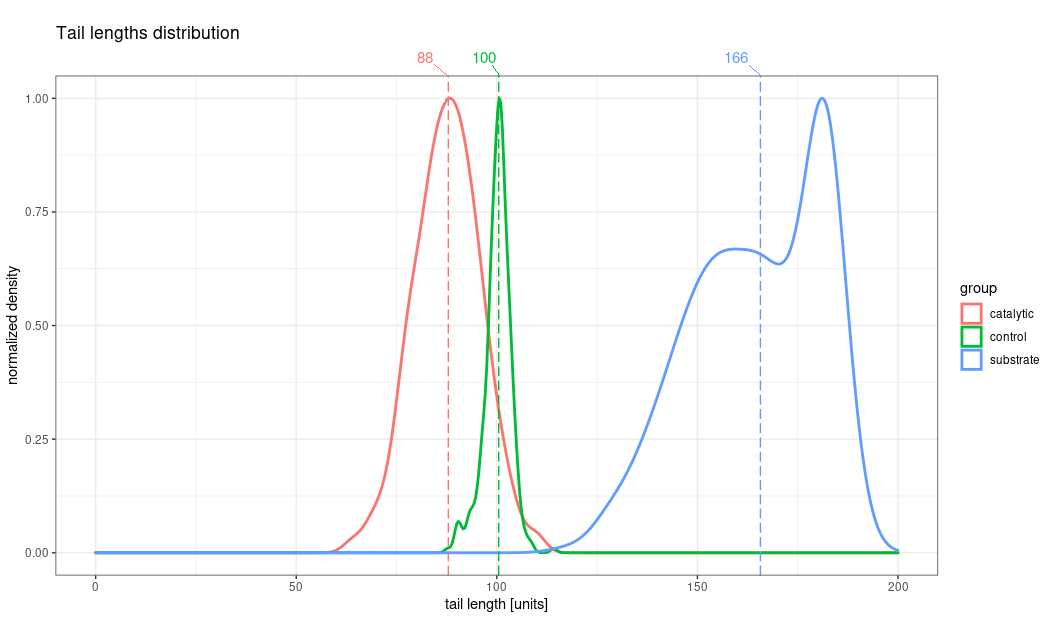
plot_distribution_with_values(tail_data, value_to_show = "median", grouping_factor = "group", title = "Tail plot", limit=200)