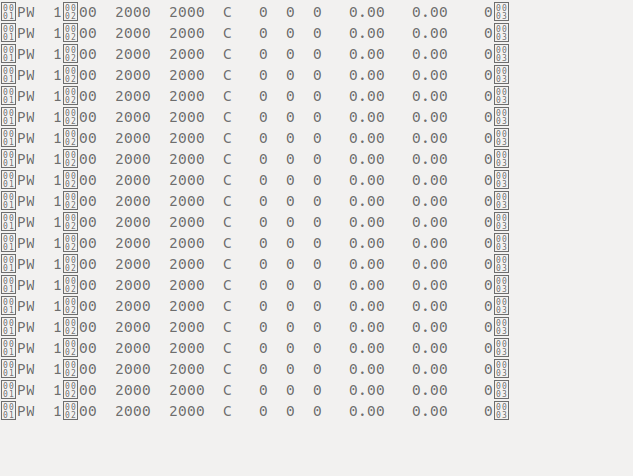
My data file contain some characters that can not be defined from keybord to set as separator. Is there anyways to read this data in proper way.
My data looks different in .txt file but when I pasted here it looks like:
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
PW 100 2000 2000 C 0 0 0 0.00 0.00 0
But I have also attached original data here data.
To read data, I simply tried by this way:
import pandas as pd
pd.read_table('data.txt',sep = '\s ')
is there better way to do that?
CodePudding user response:
You have to strip your file from invisible characters:
import pandas as pd
import io
import re
with open('pwd_data.txt') as fp:
buffer = io.StringIO(re.sub('[\01-\03]', '', fp.read()))
df = pd.read_table(buffer, header=None, sep='\s ')
Output:
>>> df
0 1 2 3 4 5 6 7 8 9 10
0 PW 100 2000 2000 C 0 0 0 0.0 0.0 0
1 PW 100 2000 2000 C 0 0 0 0.0 0.0 0
2 PW 100 2000 2000 C 0 0 0 0.0 0.0 0
...
32 PW 100 2000 2000 C 0 0 0 0.0 0.0 0
33 PW 100 2000 2000 C 0 0 0 0.0 0.0 0
34 PW 100 2000 2000 C 0 0 0 0.0 0.0 0
>>> df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 35 entries, 0 to 34
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 0 35 non-null object
1 1 35 non-null int64
2 2 35 non-null int64
3 3 35 non-null int64
4 4 35 non-null object
5 5 35 non-null int64
6 6 35 non-null int64
7 7 35 non-null int64
8 8 35 non-null float64
9 9 35 non-null float64
10 10 35 non-null int64
dtypes: float64(2), int64(7), object(2)
memory usage: 3.1 KB
CodePudding user response:
Try changing your sep to r'[\s \x00-\x19]'
pd.read_table('data.txt',sep = r'[\s \x00-\x19]')