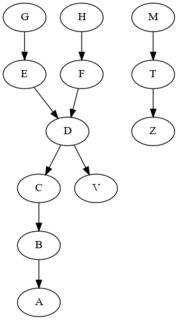
I'm trying to extract a pattern from a dataset in order to identify specific element in dataframe. Here's a sample dataset that I'm using:
import pandas as pd
import numpy as np
df = pd.DataFrame(np.array([['A', 'B'], ['B', 'C'], ['C', 'D'],['D', 'E'], ['D', 'F'], ['E', 'G'],['F', 'H'], ['Z', 'T'], ['T', 'M'],['V','D']]),
columns=['to', 'from'])
| to | from |
|---|---|
| A | B |
| B | C |
| C | D |
| D | E |
| D | F |
| E | G |
| F | H |
| Z | T |
| T | M |
| V | D |
The table represents a series of links between elements. The meaning is that A is calculated from B, B is calculated from C, C is calculated from D, D is calculated from E and F, etc.
I'm trying to identify all the input elements given a specific element. Here's my try:
obj = 'A'
target = []
flag = True
while len(df[df['to'] == obj])>0:
for i in range(0, len(df[df['to'] == obj])):
for m in range(0, len(df)):
if obj == df.loc[m,'to']:
target.append(df.loc[m,'from'])
obj = df.loc[m,'from']
print(obj)
target = target[-1]
print(target)
With that code, I obtain G as the only input element but i the main problem is that the element D is calculated from 2 elements and i can't manage to obtain G and H as input.
I hope I have been clear and comprehensive and I thank in advance anyone who can help me.
Thank you,
Giammaria
CodePudding user response:
What you want to achieve is a graph problem.
You want to find the root(s) of the subtree that contains your element.
getting all roots per element
For this you need to split the graph in disjoint subgraphs and to find the root(s) of each graph. Then you can use a dictionary comprehension to build the matches:
groups = list(nx.weakly_connected_components(G))
# [{'A', 'B', 'C', 'D', 'E', 'F', 'G', 'H', 'V'}, {'M', 'T', 'Z'}]
subgraphs = [G.subgraph(c) for c in groups]
roots = [{node for node,degree in list(g.in_degree()) if degree == 0}
for g in subgraphs]
# [{'G', 'H'}, {'M'}]
out = pd.Series({node: nx.ancestors(g, node).intersection(r)
for s,g,r in zip(groups, subgraphs, roots) for node in s},
name='root')
df.merge(out, left_on='to', right_index=True)
output:
to from root
0 A B {G, H}
1 B C {G, H}
2 C D {G, H}
3 D E {G, H}
4 D F {G, H}
5 E G {G}
6 F H {H}
7 Z T {M}
8 T M {M}
9 V D {G, H}
CodePudding user response:
This is a graph problem. You can implement your own graph traversal algorithms if you want the exercise, but otherwise it's best to use a standard library for such tasks.
The networkx library is very easy to use and has lots of convenient functions for things like this.
import pandas as pd
import numpy as np
import networkx as nx
df = pd.DataFrame(np.array([['A', 'B'], ['B', 'C'], ['C', 'D'],['D', 'E'], ['D', 'F'], ['E', 'G'],['F', 'H'], ['Z', 'T'], ['T', 'M'],['V','D']]),
columns=['to', 'from'])
g = nx.DiGraph()
g.add_edges_from(df[['from', 'to']].values)
print(nx.ancestors(g, 'D'))
The code above prints:
{'F', 'G', 'H', 'E'}