I have a CSV with many values. Among them are times stored like this:
1:34.434
Using readr, I form them into a tibble, and use dplyr to cut out all the unnecessary rows and columns.
lapData <- read_csv("sampledata/sampledata.csv")
lapData <- dplyr::select(lapData,
Driver, Lap, Penalty, 'Lap Time', 'Lap type',
'Pressure FR', 'Pressure FL', 'Pressure RR', 'Pressure RL',
'Temp FR', 'Temp FL', 'Temp RR', 'Temp RL',
'Road Temp')
lapData <- dplyr::filter(lapData, Driver == "[personal info]")
lapData <- dplyr::filter(lapData, is.na(Penalty))
lapData <- dplyr::filter(lapData, Lap > 1)
I then use print(ggplot(data = lapData, mapping = aes(Lap, 'Lap Time')) geom_line()) to print the data, and it looks like this:
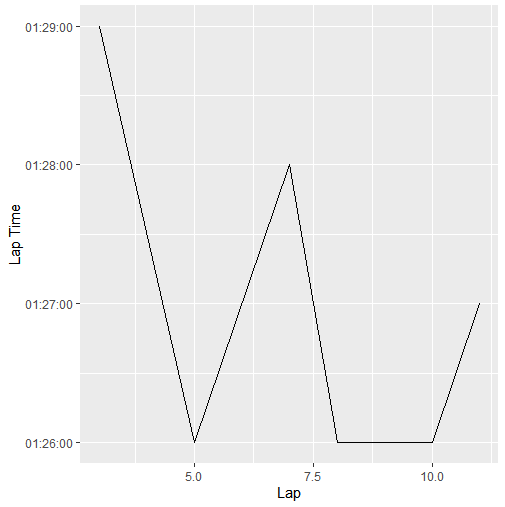
If it isn't clear, the vertices of the graph line are rounded to the seconds value instead of using the full milisecond precision, which I would like to have.
If I then print(lapData), I see the following entries under 'Lap Time':
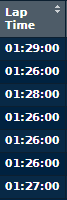
I did some research, and found 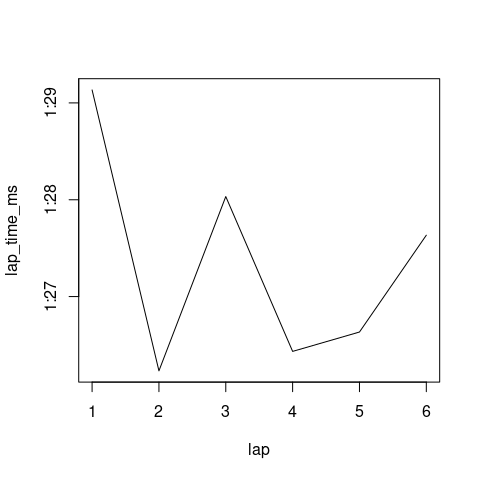
This should also be possible with ggplot2 package.
Data:
dat <- read.table(header=TRUE, text='lap_time
1:29.134
1:26.233
1:28.033
1:26.434
1:26.634
1:27.634
') |> transform(lap=1:6, x=rnorm(6))
write.csv(dat, 'foo.csv', row.names=FALSE)
