I am fitting a stateful RNN with embedding layer to perform binary classification. I am having some confusion with the batch_size and batch_shape needed in the function APIs.
xtrain_padded.shape = (9600, 1403); xtest_padded.shape = (2400, 1403); ytest.shape = (2400,)
input_dim = size of tokenizer word dictionary
output_dim = 100 from GloVe_100d embeddings
number of SimpleRNN layer units = 200
h0: initial hidden states sampled from random uniform.
h0 object has the same shape as RNN layer hidden states obtained when return_state = True.
The model structure:
batch_size = 2400 # highest common factor of xtrain and xtest
inp= Input(batch_shape= (batch_size, input_length), name= 'input')
emb_out= Embedding(input_dim, output_dim, input_length= input_length,
weights= [Emat], trainable= False, name= 'embedding')(inp)
rnn= SimpleRNN(200, return_sequences= True, return_state= True, stateful= True,
batch_size= (batch_size, input_length, 100), name= 'simpleRNN')
h_0 = tf.random.uniform((batch_size, input_length, 200))
rnn_out, rnn_state = rnn(emb_out, initial_state=h0)
mod_out= Dense(1, activation= 'sigmoid')(rnn_out)
# Extract the y_t's and h_t's:
model = Model(inputs=inp, outputs=[mod_out, rnn_out])
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['acc'])
model.summary()
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) [(2400, 1403)] 0
_________________________________________________________________
embedding (Embedding) (2400, 1403, 100) 4348900
_________________________________________________________________
simpleRNN (SimpleRNN) [(2400, 1403, 200), (2400 60200
_________________________________________________________________
dense_3 (Dense) (2400, 1403, 1) 201
No issue when I fit the test data to model using the model API:
mod_out_allsteps, rnn_ht= model(xte_pad) # Same as the 2 items from model.predict(xte_pad)
print(mod_out_allsteps.shape, rnn_ht.shape)
>> (2400, 1403, 1) (2400, 1403, 200)
However it raised a ValueError regarding unequal dimensions when I use model.fit.
model.fit(xte_pad, yte, epochs =1, batch_size = batch_size, verbose = 1)
>>
ValueError: Dimensions must be equal, but are 2400 and 1403 for '{{node binary_crossentropy_1/mul}} = Mul[T=DT_FLOAT](binary_crossentropy_1/Cast, binary_crossentropy_1/Log)' with input shapes: [2400,1], [2400,1403,200].
The error seems to suggest the model has confused the returned hidden states rnn_ht shaped [2400,1403,200] with something else when fitting the data. However I am going to need these states for computing the gradients on the initial hidden states i.e.
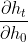 for t = 1,..., 1403.
for t = 1,..., 1403.
I am confused with the dimensions in stateful RNNs:
- If stateful = True, are we constructing the model based on one mini-batch?
i.e. the first index in Output Shape of each layer will be the batch_size? - What is the batch_shape to be set in the first layer (Input)? Have I set it right?
Thank you in advance for helping with the error and my confusion!
Update:
batch_size = 2400 # highest common factor of xtrain and xtest
input_length = 1403
output_dim = 100
inp= tf.keras.layers.Input(batch_shape= (batch_size, input_length), name= 'input')
emb_out= tf.keras.layers.Embedding(500, output_dim, input_length= input_length, trainable= False, name= 'embedding')(inp)
rnn= tf.keras.layers.SimpleRNN(200, return_sequences= True, return_state= False, stateful= True,
batch_size= (batch_size, input_length, 100), name= 'simpleRNN')
rnn_ht= rnn(emb_out) # hidden states at all steps
print(rnn_ht.shape)
>>>
(2400, 1403, 200)
mod_out= Dense(1, activation= 'sigmoid')(Flatten()(rnn_ht))
# Extract the y_t's and h_t's:
model = tf.keras.Model(inputs=inp, outputs=[mod_out, rnn_ht])
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['acc'])
model.summary()
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) [(2400, 1403)] 0
_________________________________________________________________
embedding (Embedding) (2400, 1403, 100) 50000
_________________________________________________________________
simpleRNN (SimpleRNN) (2400, 1403, 200) 60200
_________________________________________________________________
flatten_4 (Flatten) (2400, 280600) 0
_________________________________________________________________
dense_4 (Dense) (2400, 1) 280601
mod_out_allsteps, rnn_ht= model_ht(xte_pad)
print(mod_out_allsteps.shape, rnn_ht.shape)
>>>
(2400, 1) (2400, 1403, 200)
But the error with ```model.fit``` persists.
CodePudding user response:
Look at the last layer in your model summary. Since you set the parameter return_sequences to True in the RNN layer, you are getting a sequence with the same number of time steps as your input and an output space of 200 for each timestep, hence the shape (2400, 1403, 200), where 2400 is the batch size. If you set this parameter to False, everything should work, since your labels have the shape (2400, 1).
Working example:
import tensorflow as tf
batch_size = 2400 # highest common factor of xtrain and xtest
input_length = 1403
output_dim = 100
inp= tf.keras.layers.Input(batch_shape= (batch_size, input_length), name= 'input')
emb_out= tf.keras.layers.Embedding(500, output_dim, input_length= input_length, trainable= False, name= 'embedding')(inp)
rnn= tf.keras.layers.SimpleRNN(200, return_sequences= False, return_state= True, stateful= True,
batch_size= (batch_size, input_length, 100), name= 'simpleRNN')
rnn_out, rnn_state = rnn(emb_out)
mod_out= tf.keras.layers.Dense(1, activation= 'sigmoid')(rnn_out)
# Extract the y_t's and h_t's:
model = tf.keras.Model(inputs=inp, outputs=[mod_out, rnn_out])
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['acc'])
model.summary()
where the first output is your binary decision.
Update 1: with Flatten:
import tensorflow as tf
batch_size = 2400 # highest common factor of xtrain and xtest
input_length = 1403
output_dim = 100
inp= tf.keras.layers.Input(batch_shape= (batch_size, input_length), name= 'input')
emb_out= tf.keras.layers.Embedding(500, output_dim, input_length= input_length, trainable= False, name= 'embedding')(inp)
rnn= tf.keras.layers.SimpleRNN(200, return_sequences= True, return_state= True, stateful= True,
batch_size= (batch_size, input_length, 100), name= 'simpleRNN')
rnn_out, rnn_state = rnn(emb_out)
mod_out= tf.keras.layers.Dense(1, activation= 'sigmoid')(tf.keras.layers.Flatten()(rnn_out))
# Extract the y_t's and h_t's:
model = tf.keras.Model(inputs=inp, outputs=[mod_out, rnn_out])
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['acc'])
model.summary()
