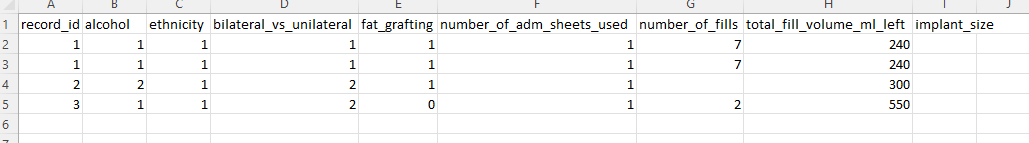
I have data like this:
df<-structure(list(record_id = c(1, 2, 4), alcohol = c(1, 2, 1),
ethnicity = c(1, 1, 1), bilateral_vs_unilateral = c(1, 2,
2), fat_grafting = c(1, 1, 0), number_of_adm_sheets_used = c(1,
NA, NA), number_of_adm_sheets_used_2 = c(1, 1, 1), number_of_fills = c(7,
NA, NA), number_of_fills_2 = c(7, NA, 2), total_fill_volume_ml_left = c(240,
NA, NA), total_volume_ml = c(240, 300, 550), implant_size_l = c(NA_real_,
NA_real_, NA_real_), implant_size_l_2 = c(NA_real_, NA_real_,
NA_real_)), row.names = c(NA, -3L), class = c("tbl_df", "tbl",
"data.frame"))
It is info about patients with each row representing a patient that underwent breast surgery.
I'd like to change it into each row representing a particular breast (of the two). There are several variables, everything from 'number_of_adm_sheets_used' to 'implant_size_l_2' that have a column for each side. I'd like to change those to represent either. An example is 'number_of_adm_sheets_used' stood for on the left side, and 'number_of_adm_sheets_used_2' was on the right side. I'd like to combine them to become one column of sheets used that was for either side.
My expected output would look like:
I figure its some variant of pivot_longer but I'm having trouble with a few aspects:
- the real data has 68 columns
- I only need a duplicate row if the column "bilateral_vs_unilateral" is a "1" (meaning bilateral)
- The way I've used pivot_longer before, you'd say "cols" and pick a big range, I'm not sure how to stack pairs of columns, if that makes sense.
- Luckily, despite having 68 other columns, all of the "trouble" columns are shown below. Pairing 'number_of_adm_sheets_used' with 'number_of_adm_sheets_used_2' 'number_of_fills' with 'number_of_fills_2' 'total_fill_volume_ml_left' with 'total_volume_ml' and 'implant_size_1' with 'implant_size_1_2'
Thank you
CodePudding user response:
Here is one possibility, if I'm understanding the issue correctly.
# Make long format
df.long <- df %>%
pivot_longer(cols = -record_id) %>%
mutate(subject = ifelse(str_sub(name, -2, -1) == "_2", "breast 2", NA),
name = str_remove(name, "_2")) %>%
group_by(record_id, name) %>%
mutate(subject = case_when(
subject == "breast 2" ~ subject,
n() == 2 ~ "breast 1",
n() == 1 ~ "patient"
)) %>%
ungroup()
# statistics regarding the patient
patient <- df.long %>%
filter(subject == "patient") %>%
pivot_wider(names_from = name, values_from = value) %>%
select(-subject)
# statistics regarding each breast
breasts <- df.long %>%
filter(str_detect(subject, "breast")) %>%
pivot_wider(names_from = name, values_from = value)
# merge the two data.frames
patient %>%
inner_join(breasts) %>%
select(record_id, subject, everything())