I'm trying to use scipy minimize to estimate a parameter from an ODE system, which is pretty straightforward, however the methods I used aren't returning values anywhere near what the value should be. My parameter beta, should have a value estimated to be around 0.42. I am sure that this method is correct, so i can't understand why the estimates are so off
import numpy as np
from scipy.integrate import odeint
from scipy.optimize import minimize
from scipy.optimize import minimize_scalar
import pandas as pd
from scipy.optimize import leastsq
t = np.linspace(0, 77, 77 1)
d = {'Week': [t[7],t[14],t[21],t[28],t[35],t[42],t[49],t[56],t[63],t[70],t[77]],
'incidence': [206.1705794,2813.420201,11827.9453,30497.58655,10757.66954,
7071.878779,3046.752723,1314.222882,765.9763902,201.3800578,109.8982006]}
df = pd.DataFrame(data=d)
def peak_infections(beta, df):
# Weeks for which the ODE system will be solved
#weeks = df.Week.to_numpy()
# Total population, N.
N = 100000
# Initial number of infected and recovered individuals, I0 and R0.
I0, R0 = 10, 0
# Everyone else, S0, is susceptible to infection initially.
S0 = N - I0 - R0
J0 = I0
# Contact rate, beta, and mean recovery rate, gamma, (in 1/days).
#reproductive no. R zero is beta/gamma
gamma = 1/6 #rate should be in weeks now
# A grid of time points
times = np.arange(7,84,7)
# The SIR model differential equations.
def deriv(y, times, N, beta, gamma):
S, I, R, J = y
dS = ((-beta * S * I) / N)
dI = ((beta * S * I) / N) - (gamma * I)
dR = (gamma * I)
dJ = ((beta * S * I) / N) #incidence
return dS, dI, dR, dJ
# Initial conditions are S0, I0, R0
# Integrate the SIR equations over the time grid, t.
solve = odeint(deriv, (S0, I0, R0, J0), times, args=(N, beta, gamma))
S, I, R, J = solve.T
return I/N
def residual(x, df):
# Total population, N.
N = 100000
incidence = df.incidence.to_numpy()/N
return np.sum((peak_infections(x,df) - incidence) ** 2)
x0 = 0.5
res = minimize(residual, x0, args=(df), method="Nelder-Mead", options={'fatol':1e-04}).x
print(res)
best = leastsq(residual, x0,args=(df))
print(best) #tried this using leastsq too
results = minimize_scalar(residual,(0.4, 0.5),args=(df))
print(results)
results['fun']
As you can see I have used minimize, minimize_scalar and even leastsq. They all returns values such as 0.723. Where have I gone wrong? Is my objective function return np.sum((peak_infections(x,df) - incidence) ** 2) at least correct?
edit: i tried taking the np.max(I/N) in the peak_infections(beta, df) function however this does not return a correct estimate either
CodePudding user response:
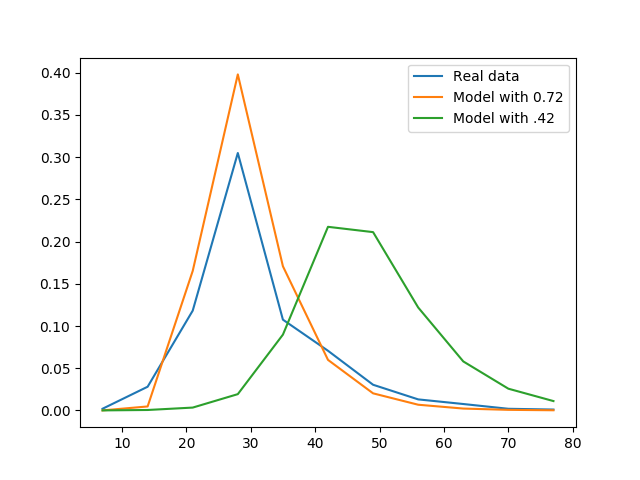
Assuming that the model is OK, the problem is that you assume that beta should be close to 0.42 when it should not. A simple visual test, plotting the measurements and the modeled data, shows that 0.72 yields much better results than 0.43. I added the following lines:
import matplotlib.pyplot as plt
plt.plot(d['Week'], df.incidence.to_numpy()/100000, label="Real data")
plt.plot(d['Week'], peak_infections(.72, df), label="Model with 0.72")
plt.plot(d['Week'], peak_infections(.42, df), label="Model with .42")
plt.legend()
which yielded the following figure:
It is clear that 0.72 is a better estimate of beta than 0.42, even without plotting the residuals.
As a side note, be careful because your code is easy to break.You define the same values in many places: N is defined inside peak_infections and residual. The days of the week are defined as d['Week'] and as times inside peak_infections. It is easy to change the value of one of this variables in one place and forget to change it in another place. Moreover, I see no use of pandas here, since you do not take profit of its features on top of numpy.