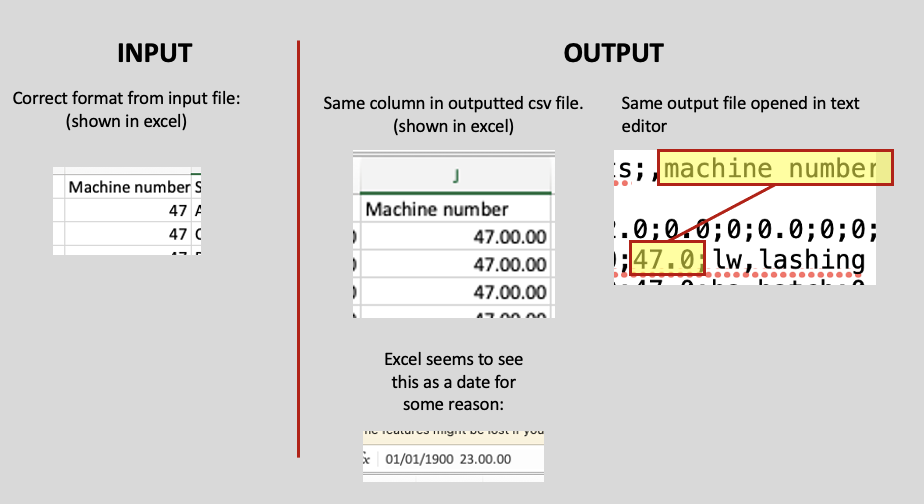
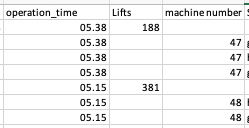
I'm trying to merge a series of xlsx files into one, which works fine. However, when I read a file, columns containing ints are transformed into floats (or dates?) when I merge and output them to csv. I have tried to visualize this in the picture. I have seen some solutions to this where dtype is used to "force" specific columns into int format. However, I do not always know the index nor the title of the column, so i need a more scalable solution.
Anyone with some thoughts on this?
Thank you in advance
#specify folder with xlsx-files
xlsFolder = "{}/system".format(directory)
dfMaster = pd.DataFrame()
#make a list of all xlsx-files in folder
xlsFolderContent = os.listdir(xlsFolder)
xlsFolderList = []
for file in xlsFolderContent:
if file[-5:] == ".xlsx":
xlsFolderList.append(file)
for xlsx in xlsFolderList:
print(xlsx)
xl = pd.ExcelFile("{}/{}".format(xlsFolder, xlsx))
for sheet in xl.sheet_names:
if "_Errors" in sheet:
print(sheet)
dfSheet = xl.parse(sheet)
dfSheet.fillna(0, inplace=True)
dfMaster = dfMaster.append(dfSheet)
print("len of dfMaster:", len(dfMaster))
dfMaster.to_csv("{}/dfMaster.csv".format(xlsFolder),sep=";")CodePudding user response:
Try to use dtype='object' as parameter of pd.read_csv or (ExcelFile.parse) to prevent Pandas to infer the data type of each column. You can also simplify your code using pathlib:
import pandas as pd
import pathlib
directory = pathlib.Path('your_path_directory')
xlsFolder = directory / 'system'
data = []
for xlsFile in xlsFolder.glob('*.xlsx'):
sheets = pd.read_excel(xlsFile, sheet_name=None, dtype='object')
for sheetname, df in sheets.items():
if '_Errors' in sheetname:
data.append(df.fillna('0'))
pd.concat(data).to_csv(xlsxFolder / dfMaster.csv, sep=';')