I have created the following plot:
From a bigger version (5 rows, 58 columns) of this df:
df <- data.frame(row.names = c("ROBERT", "FRANK", "MICHELLE", "KATE"), `1` = c(31, 87, 22, 12), `2` = c(37, 74, 33, 20), `3` = c(35, 32, 44, 14))
colnames(df) <- c("1", "2", "3")
In the following manner:
df = df %>%
rownames_to_column("Name") %>%
as.data.frame()
df <- melt(df , id.vars = 'Name', variable.name = 'ep')
ggplot(df, aes(ep,value)) geom_line(aes(colour = Name, group=Name))
The plot kind of shows what I'd like to, but it really is a mess. Does anyone have a suggestion that would help me increasing its readability? Any help is very much appreciated!
CodePudding user response:
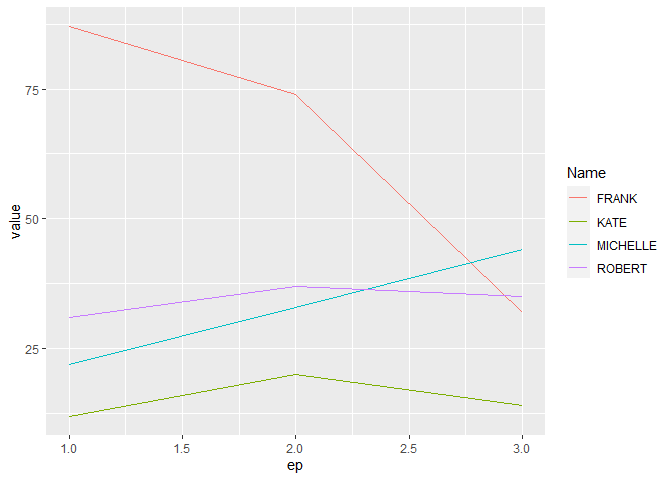
If your readability concern is just the x axis labels, then I think the main issue is that when you use reshape2::melt() the result is that the column ep is a factor which means that the x axis of your plot will show all the levels and get crowded. The solution is to convert it to numeric and then it will adjust the labels in a sensible way.
I replace your use of reshape2::melt() with tidyr::pivot_longer() which has superseded it within the {tidyverse} but your original code would still work.
library(tidyverse)
df <- structure(list(`1` = c(31, 87, 22, 12), `2` = c(37, 74, 33, 20), `3` = c(35, 32, 44, 14)), class = "data.frame", row.names = c("ROBERT", "FRANK", "MICHELLE", "KATE"))
df %>%
rownames_to_column("Name") %>%
pivot_longer(-Name, names_to = "ep") %>%
mutate(ep = as.numeric(ep)) %>%
ggplot(aes(ep, value, color = Name))
geom_line()
Simulated data:
library(tidyr)
library(dplyr)
set.seed(1)
df <- expand_grid(
Name = c("ROBERT", "FRANK", "MICHELLE", "KATE", "COREY"),
ep = 1:58
) %>%
mutate(value = sample(0:130, nrow(.), replace = TRUE))