I understand for-loops are slow in R, and the suite of apply() functions are designed to be used instead (in many cases).
However, I can't figure out how to use those functions in my situation, and advice would be greatly appreciated.
I have a list/vector of values (let's say length=10,000) and at every point, starting at the 21st value, I need to take the standard deviation of the trailing 20 values. So at 21st, I take SD of 1st-21st . At 22nd value, I take SD(2:22) and so on.
So you see I have a rolling window where I need to take the SD() of the previous 20 indices. Is there any way to accomplish this faster, without a for-loop?
CodePudding user response:
I found a solution to my question.
The zoo package has a function called "rollapply" which does exactly that: uses apply() on a rolling window basis.
CodePudding user response:
library(microbenchmark)
library(ggplot2)
# dummy vector
x <- sample(1:100, 50, replace=T)
# parameter
y <- 20 # length of each vector
z <- length(x) - y # final starting index
# benchmark
xx <-
microbenchmark(lapply = {a <- lapply( 1:z, \(i) sd(x[i:(i y)]) )}
, loop = {
b <- list()
for (i in 1:z)
{
b[[i]] <- sd(x[i:(i y)])
}
}
, times = 30
)
# plot
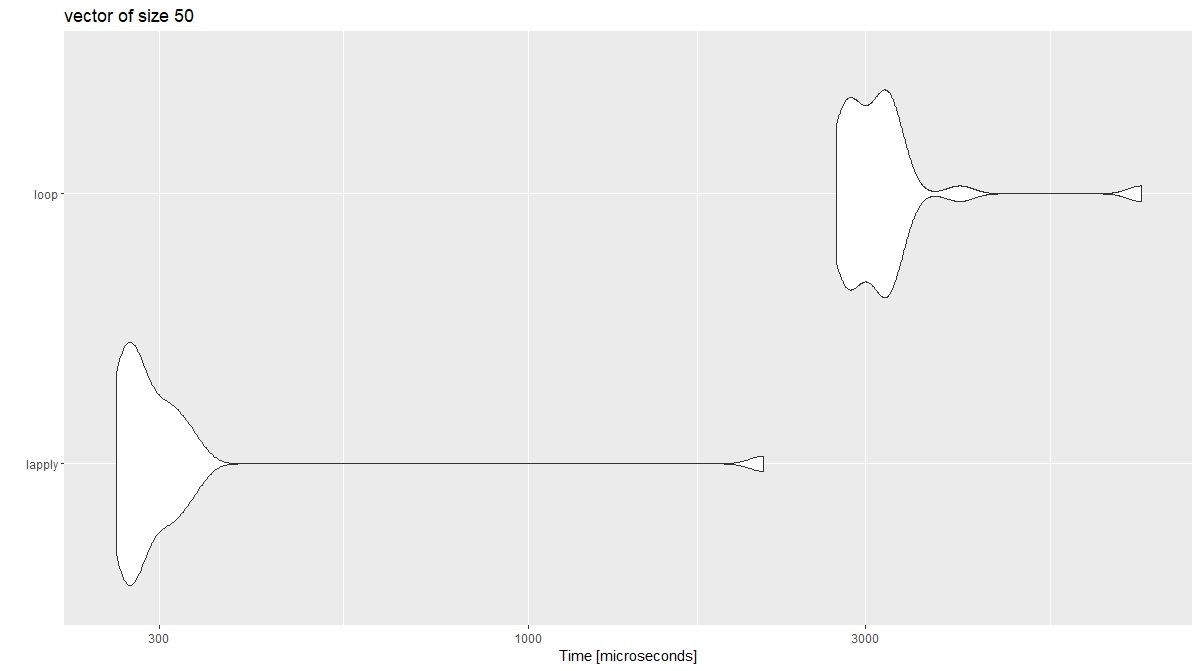
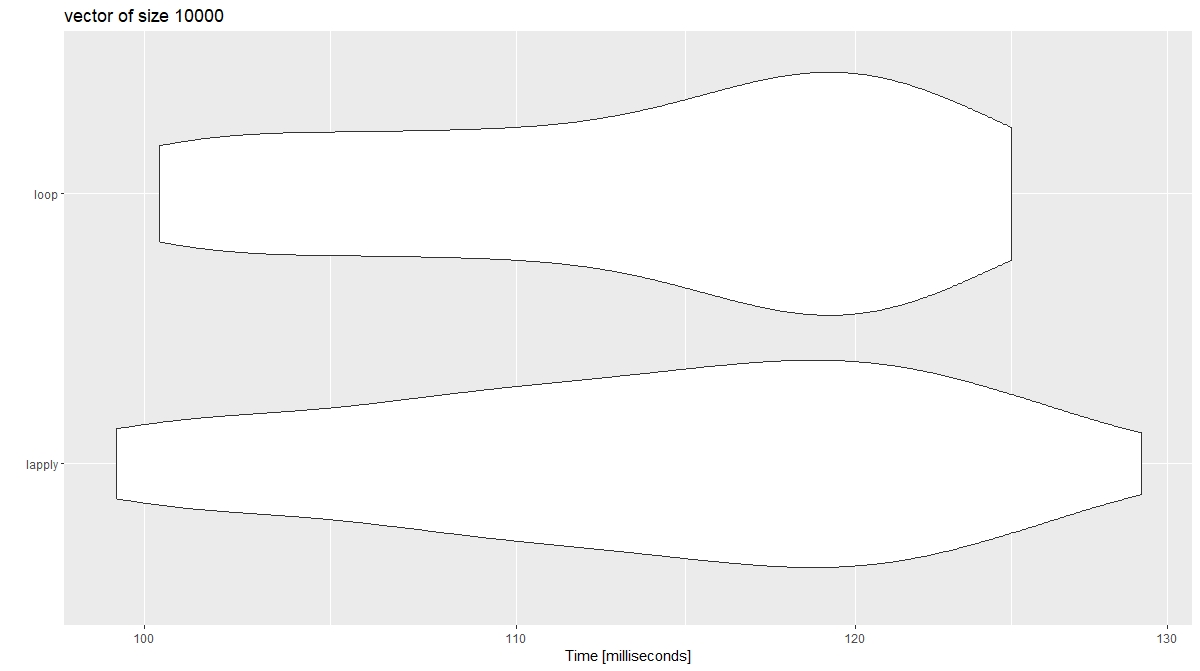
autoplot(xx)
It would appear while lapply has the speed advantage of a smaller vector, a loop should be used with longer vectors.
EDIT* Results vary from run to run with a vector of length 1e4. It would appear other larger factors are at play. I would maintain, however, loops are not slow per se as long as they are not applied incorrectly (iterating over rows).