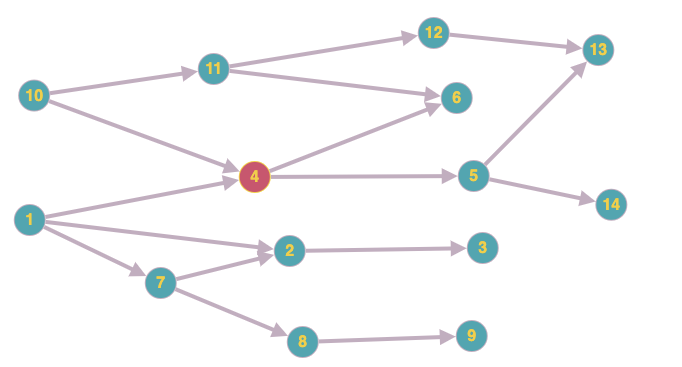
Let's assume I have this directed graph:
What's the optimal (complexity and simplicity) way to get every path that passes through a given node?
Let's say I choose node = 4 I want to get this sub-graph:
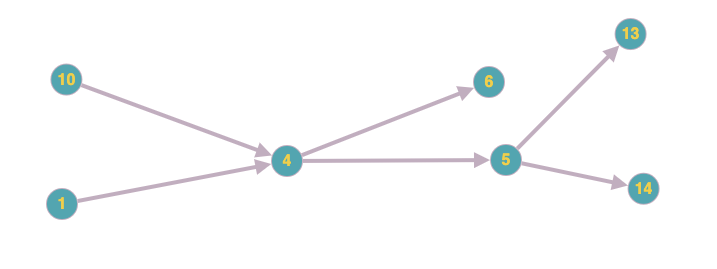 Currently I am using NetworkX python library, however I am open to choosing another one if it has this capability.
Currently I am using NetworkX python library, however I am open to choosing another one if it has this capability.
CodePudding user response:
Let me propose an "ugly" solution (and I am sure there is a better way). First, find all descendants and ancestors of the node:
desc = nx.descendants(G, 4)
anc = nx.ancestors(G, 4)
Next, find the simple paths from each ancestor to the node and from the node to each descendant:
paths = [path for p in desc for path in nx.all_simple_paths(G, 4, p)]
paths.extend([path for p in anc for path in nx.all_simple_paths(G, p, 4)])
Finally, reconstruct the digraph from the paths:
Q = nx.DiGraph()
for p in paths:
nx.add_path(Q, p)
CodePudding user response:
Here is another approach. As in the answer by DYZ, let anc denote the ancestors and desc denote the descendants of the chosen node. Let nodes = anc union desc union {chosen_node}. Iterate over edges (i, j) of the original graph, and delete an edge if either of the following holds:
iorjare not in the setnodes.iis inancandjis indesc.
A node x not in nodes cannot be on any path containing chosen_node, or else x would have to be an ancestor, a descendant, or chosen_node itself (condition 1). If (i, j) is a node from an ancestor i to descendant j, it cannot be on any path containing chosen_node because of the acyclic assumption (condition 2).
Every remaining edge is on some path containing the chosen_node. I believe you can prove it by induction on min(distance(i, chosen_node), distance(j, chosen_node)).
def remove_edges(G, node=0):
desc = nx.descendants(G, node)
anc = nx.ancestors(G, node)
nodes = anc.union(desc).union({node})
Q = nx.DiGraph()
for i, j in G.edges:
# remove edges with either source or target not in nodes
if (i not in nodes) or (j not in nodes):
continue
# remove edges immediately connecting ancestors to descendants
if (i in anc) and (j in desc):
continue
Q.add_edge(i,j)
return Q