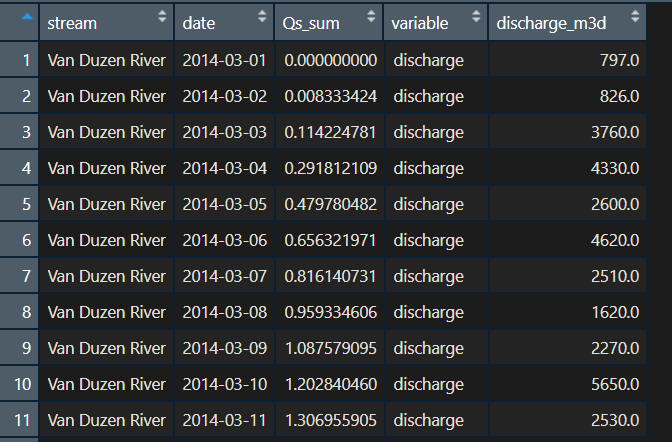
I have daily discharge data from a local stream near me. I am trying to sum and take the average of the daily data into weekly or monthly chunks so I can plot discharge_m3d(discharge) and Qs_sum(depletion) by weekly and monthly timeframes. Does anyone know how I can do this? I attached a figure of how my data frame looks.
CodePudding user response:
One way to approach this is using the lubridate and dplyr packages in the tidyverse. I assume here that your dates are year-month-day which they appear to be and that you only have one calendar year or at least no repeated months/weeks across two years.
monthly_discharge <- discharge %>%
filter(variable == "discharge") # First select just the rows that represent discharge (not clear if that's necessary here)
mutate(date = ymd(date), # convert date to a lubridate date object
month = month(date), # extract the numbered month from the date
week = week(date)) %>% # extract the numbered week in a year from the date
group_by(month, stream) %>% # group your data by month and stream
summarize(discharge_summary = mean(discharge_m3d)) # summarize your data so that each month has a single row with a single (mean) discharge value
# you can include multiple summary variables within the summarize function
This should produce a data frame with one row per month for each stream and a summary value for discharge. You could summarize by week by changing the month label in group_by to week.
CodePudding user response:
Make use of the functions week() and month() from the package lubridate to get the corresponding values for your date column. Afterwards we can find the means per week (or month):
library(dplyr)
library(lubridate)
data <- data %>% mutate(Week = week(date), Month = month(date)) %>% group_by(Week, Month) %>%
mutate(mean_Week_Qs = mean(Qs_sum)) %>% ungroup()
> head(data)
# A tibble: 6 x 6
date discharge_m3d Qs_sum Week Month mean_Week_Qs
<date> <dbl> <dbl> <int> <int> <dbl>
1 2014-03-01 797 0 9 3 0.0409
2 2014-03-02 826 0.00833 9 3 0.0409
3 2014-03-03 3760 0.114 9 3 0.0409
4 2014-03-04 4330 0.292 10 3 0.785
5 2014-03-05 2600 0.480 10 3 0.785
6 2014-03-06 4620 0.656 10 3 0.785
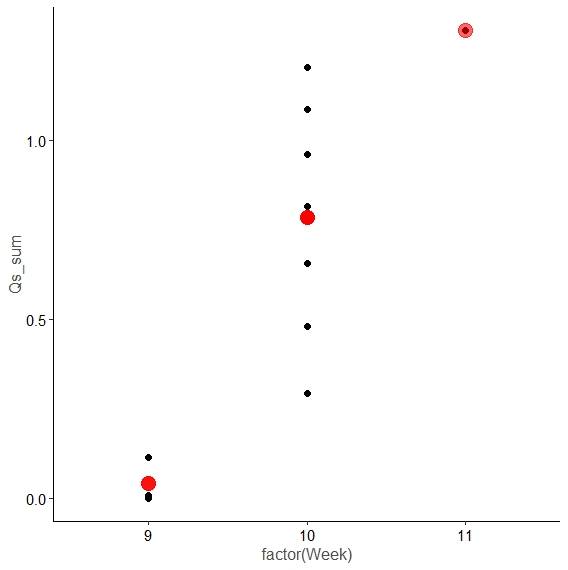
Now we can plot, for example Qs_sum per week, and add the mean as a red dot:
ggplot(data, aes(factor(Week), Qs_sum))
geom_point(size = 2)
geom_point(aes(factor(Week), mean_Week_Qs), color = "red", size = 5, alpha = 0.6)
Data
data <- structure(list(date = structure(16130:16140, class = "Date"),
discharge_m3d = c(797, 826, 3760, 4330, 2600, 4620, 2510,
1620, 2270, 5650, 2530), Qs_sum = c(0, 0.00833424, 0.114224781,
0.291812109, 0.479780482, 0.656321971, 0.816140731, 0.959334606,
1.087579095, 1.20284046, 1.30695595), Week = c(9L, 9L, 9L,
10L, 10L, 10L, 10L, 10L, 10L, 10L, 11L), Month = c(3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L)), row.names = c(NA, -11L
), class = c("tbl_df", "tbl", "data.frame"))
CodePudding user response:
People often use floor_date() from lubridate for these purposes. You can floor to a unit of month or week and then group by the resulting date column. Then you can use summarize() to compute the monthly or weekly sums/averages. From there you can use your plotting library of choice to visualize the result (like ggplot2, not shown).
This works even if you have more than one year of data (i.e. where the month or week number might repeat).
library(dplyr)
library(lubridate)
set.seed(123)
df <- tibble(
date = seq(
from = as.Date("2014-03-01"),
to = as.Date("2016-12-31"),
by = 1
),
Qs_sum = runif(length(date)),
discharge_m3d = runif(length(date))
)
df
#> # A tibble: 1,037 × 3
#> date Qs_sum discharge_m3d
#> <date> <dbl> <dbl>
#> 1 2014-03-01 0.288 0.560
#> 2 2014-03-02 0.788 0.427
#> 3 2014-03-03 0.409 0.448
#> 4 2014-03-04 0.883 0.833
#> 5 2014-03-05 0.940 0.720
#> 6 2014-03-06 0.0456 0.457
#> 7 2014-03-07 0.528 0.521
#> 8 2014-03-08 0.892 0.242
#> 9 2014-03-09 0.551 0.0759
#> 10 2014-03-10 0.457 0.391
#> # … with 1,027 more rows
df %>%
mutate(date = floor_date(date, unit = "month")) %>%
group_by(date) %>%
summarise(
n = n(),
qs_total = sum(Qs_sum),
qs_average = mean(Qs_sum),
discharge_total = sum(discharge_m3d),
discharge_average = mean(discharge_m3d),
.groups = "drop"
)
#> # A tibble: 34 × 6
#> date n qs_total qs_average discharge_total discharge_average
#> <date> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 2014-03-01 31 18.1 0.585 15.3 0.494
#> 2 2014-04-01 30 12.9 0.429 15.2 0.507
#> 3 2014-05-01 31 15.5 0.500 15.3 0.493
#> 4 2014-06-01 30 15.8 0.525 16.3 0.542
#> 5 2014-07-01 31 15.1 0.487 13.9 0.449
#> 6 2014-08-01 31 14.8 0.478 16.2 0.522
#> 7 2014-09-01 30 15.3 0.511 13.1 0.436
#> 8 2014-10-01 31 15.6 0.504 14.7 0.475
#> 9 2014-11-01 30 16.0 0.532 15.1 0.502
#> 10 2014-12-01 31 14.2 0.458 15.5 0.502
#> # … with 24 more rows
# Assert that the "start of the week" is Sunday.
# So groups are made of data from [Sunday -> Monday]
sunday <- 7L
df %>%
mutate(date = floor_date(date, unit = "week", week_start = sunday)) %>%
group_by(date) %>%
summarise(
n = n(),
qs_total = sum(Qs_sum),
qs_average = mean(Qs_sum),
discharge_total = sum(discharge_m3d),
discharge_average = mean(discharge_m3d),
.groups = "drop"
)
#> # A tibble: 149 × 6
#> date n qs_total qs_average discharge_total discharge_average
#> <date> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 2014-02-23 1 0.288 0.288 0.560 0.560
#> 2 2014-03-02 7 4.49 0.641 3.65 0.521
#> 3 2014-03-09 7 3.77 0.539 3.88 0.554
#> 4 2014-03-16 7 4.05 0.579 3.45 0.493
#> 5 2014-03-23 7 4.43 0.632 3.08 0.440
#> 6 2014-03-30 7 4.00 0.572 4.74 0.677
#> 7 2014-04-06 7 2.50 0.357 3.15 0.449
#> 8 2014-04-13 7 2.48 0.355 2.44 0.349
#> 9 2014-04-20 7 2.30 0.329 2.45 0.349
#> 10 2014-04-27 7 3.44 0.492 4.40 0.629
#> # … with 139 more rows
Created on 2022-04-13 by the reprex package (v2.0.1)