I have data as follows:
Previous solution
library(dplyr)
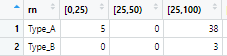
dat_in <- structure(list(rn = c("Type_A", "Type_B"
), `[0,25)` = c(5L, 0L), `[25,50)` = c(0L, 0L), `[25,100)` = c(38L,
3L), `[50,100)` = c(0L, 0L), `[100,250)` = c(43L, 5L), `[100,500)` = c(0L,
0L), `[250,500)` = c(27L, 12L), `[500,1000)` = c(44L, 0L), `[1000,1500)` = c(0L,
0L), `[1500,3000)` = c(0L, 0L), `[500,1000000]` = c(0L, 53L),
`[1000,1000000]` = c(20L, 0L), `[3000,1000000]` = c(0L, 0L
), Sum_bin = c(177, 73), strata = list(c(0, 25, 100, 250,
500, 1000, 1e 06), c(0, 25, 100, 250, 500, 1e 06))), row.names = c(NA,
-2L), class = c("data.table", "data.frame"))
New issue
Regrettably this solution was not robust enough for the data (because for example the [0,25) strata actually had zero frequencies for type_B).
What I would like to try is to create a list of values in the freq column, based on the strata column.
[[1]]
[1] 0 25 100 250 500 1000 1000000
[[2]]
[1] 0 25 100 250 500 1000000
So for the first row, the values in [0,25), [25,100), [100,250), [250,500), [500,1000) and [1000,1000000) should be collected.
But the following row has different strata values
For the second row the values [0,25), [25,100), [100,250), [250,500), and [500,1000000) should be collected.
I am having a hard time thinking of an approach to do this. Can anyone suggest a good approach?
Desired output:
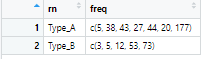
dat_out <- structure(list(rn = c("Type_A", "Type_B"), freq = list(c(5, 38,
43, 27, 44, 20, 177), c(0, 3, 5, 12, 53, 73))), class = c("rowwise_df",
"tbl_df", "tbl", "data.frame"), row.names = c(NA, -2L), groups = structure(list(
.rows = structure(list(1L, 2L), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), row.names = c(NA, -2L), class = c("tbl_df",
"tbl", "data.frame")))
CodePudding user response:
Maybe this?
library(tidyverse)
dat_in %>%
as_tibble() %>%
dplyr::select(where(~ any(unlist(.) !=0))) %>%
select(-Sum_bin, -strata) %>%
nest(freq = -rn)
which gives:
rn freq
<chr> <list>
1 Type_A <tibble [1 x 7]>
2 Type_B <tibble [1 x 7]>
Or this:
library(tidyverse)
dat_in %>%
as_tibble() %>%
dplyr::select(where(~ any(unlist(.) !=0))) %>%
select(-Sum_bin, -strata) %>%
unite(new_var, -rn, sep = ", ", remove = TRUE) %>%
nest(freq = -rn)
which gives this:
rn freq
<chr> <list>
1 Type_A <tibble [1 x 1]>
2 Type_B <tibble [1 x 1]>
CodePudding user response:
dat_in %>%
pivot_longer(-c(rn, strata)) %>%
extract(name, c('lower', 'upper'), '(\\d ),(\\d )', convert = TRUE) %>%
group_by(rn) %>%
filter(lower%in%strata[[1]] & upper %in% strata[[1]]) %>%
group_by(upper,.add = TRUE) %>%
summarise(freq = sum(value), .groups = 'drop_last') %>%
group_modify(~add_row(.,freq = sum(.$freq))) %>%
summarise(freq = list(freq))
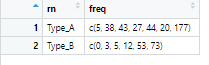
# A tibble: 2 x 2
rn freq
<chr> <list>
1 Type_A <dbl [7]>
2 Type_B <dbl [6]>
with the freq column:
[[1]]
[1] 5 38 43 27 44 20 177
[[2]]
[1] 0 3 5 12 53 73