I have several data frames that have the same columns names, and ID
, the following to are the start from and end to of a range and group label from each of them.
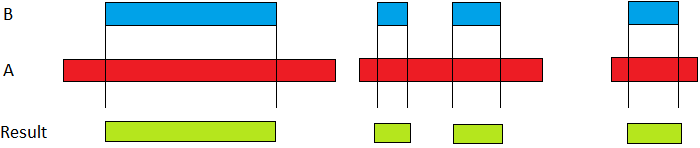
What I want is to find which values offrom and to from one of the data frames are included in the range of the other one. I leave an example picture to ilustrate what I want to achieve (no graph is need for the moment)
I thought I could accomplish this using between() of the dplyr package but no. This could be accomplish using if between() returns true then return the maximum value of from and the minimum value of to between the data frames.
I leave example data frames and the results I'm willing to obtain.
a <- data.frame(ID = c(1,1,1,2,2,2,3,3,3),from=c(1,500,1000,1,500,1000,1,500,1000),
to=c(400,900,1400,400,900,1400,400,900,1400),group=rep("a",9))
b <- data.frame(ID = c(1,1,1,2,2,2,3,3,3),from=c(300,1200,1900,1400,2800,3700,1300,2500,3500),
to=c(500,1500,2000,2500,3000,3900,1400,2800,3900),group=rep("b",9))
results <- data.frame(ID = c(1,1,1,2,3),from=c(300,500,1200,1400,1300),
to=c(400,500,1400,1400,1400),group=rep("a, b",5))
I tried using this function which will return me the values when there is a match but it doesn't return me the range shared between them
f <- function(vec, id) {
if(length(.x <- which(vec >= a$from & vec <= a$to & id == a$ID))) .x else NA
}
b$fromA <- a$from[mapply(f, b$from, b$ID)]
b$toA <- a$to[mapply(f, b$to, b$ID)]
CodePudding user response:
We can play with the idea that the starting and ending points are in different columns and the ranges for the same group (a and b) do not overlap. This is my solution. I have called 'point_1' and 'point_2' your mutated 'from' and 'to' for clarity.
You can bind the two dataframes and compare the from col with the previous value lag(from) to see if the actual value is smaller. Also you compare the previous lag(to) to the actual to col to see if the max value of the range overlap the previous range or not.
Important, these operations do not distinguish if the two rows they are comparing are from the same group (a or b). Therefore, filtering the NAs in point_1 (the new mutated 'from' column) you will remove wrong mutated values.
Also, note that I assume that, for example, a range in 'a' cannot overlap two rows in 'b'. In your 'results' table that doesn't happen but you should check that in your dataframes.
res = rbind(a,b) %>% # Bind by rows
arrange(ID,from) %>% # arrange by ID and starting point (from)
group_by(ID) %>% # perform the following operations grouped by IDs
# Here is the trick. If the ranges for the same ID and group (i.e. 1,a) do
# not overlap, when you mutate the following cols the result will be NA for
# point_1.
mutate(point_1 = ifelse(from <= lag(to), from, NA),
point_2 = ifelse(lag(to)>=to, to, lag(to)),
groups = paste(lag(group), group, sep = ',')) %>%
filter(! is.na(point_1)) %>% # remove NAs in from
select(ID,point_1, point_2, groups) # get the result dataframe
If you play a bit with the code, not using the filter() and select() you will see how that's work.
> res
# A tibble: 5 x 4
# Groups: ID [3]
ID point_1 point_2 groups
<dbl> <dbl> <dbl> <chr>
1 1 300 400 a,b
2 1 500 500 b,a
3 1 1200 1400 a,b
4 2 1400 1400 a,b
5 3 1300 1400 a,b