The original data is as below:
ISIDOR <- structure(list(Pos_heliaphen = c("W30", "X41", "Y27", "Z24",
"Y27", "W30", "W30", "X41", "Y27", "W30", "X41", "Z40", "Z99"
), traitement = c("WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW",
"WW", "WW", "WW", "WW", "WW"), Variete = c("Isidor", "Isidor",
"Isidor", "Isidor", "Isidor", "Isidor", "Isidor", "Isidor", "Isidor",
"Isidor", "Isidor", "Isidor", "Cali"), FTSW_apres_arros = c(0.462837958498518,
0.400045032939416, 0.352560790392534, 0.377856799586057, 0.170933345859364,
0.315689846065931, 0.116825600914318, 0.0332444780173884, 0.00966070114456602,
0.0871102539376406, 0.0107280083093036, 0.195548432729584, 1),
NLE = c(0.903498791068124, 0.954670066942938, 0.970762905436272,
0.873838605282389, 0.647875257025359, 0.53056603773585, 0.0384548155916796,
0.0470924009989314, 0.00403163281128882, 0.193696514297641,
0.0718450645564359, 0.295346695941639, 1)), class = c("tbl_df",
"tbl", "data.frame"), row.names = c(NA, -13L))
Here is the code:
pred_df <- data.frame(FTSW_apres_arros = seq(min(ISIDOR$FTSW_apres_arros),
max(ISIDOR$FTSW_apres_arros),
length.out = 100))
pred_df$NLE <- predict(mod, newdata = pred_df)
mod = nls(NLE ~ 2/(1 exp(a*FTSW_apres_arros))-1,start = list(a=1),data = ISIDOR)
ISIDOR$pred = predict(mod,ISIDOR)
a = coef(mod)
RMSE = rmse(ISIDOR$NLE, ISIDOR$pred)
MSE = mse(ISIDOR$NLE, ISIDOR$pred)
Rsquared = summary(lm(ISIDOR$NLE~ ISIDOR$pred))$r.squared
ggplot(ISIDOR, aes(FTSW_apres_arros, NLE))
geom_point(aes(color = Variete), pch = 19, cex = 3)
geom_line(data = pred_df)
scale_color_manual(values = c("red3","blue3"))
scale_y_continuous(limits = c(0, 1.0))
scale_x_continuous(limits = c(0, 1))
labs(title = "Isidor",
y = "Expansion folliaire totale relative",
x = "FTSW",
subtitle = paste0("y = 2/(1 exp(", round(a, 3), "* x)) -1)","\n",
"R^2 = ", round(Rsquared, 3)," RMSE = ",
round(RMSE, 3), " MSE = ", round(MSE, 3)))
theme(plot.title = element_text(hjust = 0, size = 14, face = "bold",
colour = "black"),
plot.subtitle = element_text(hjust = 0,size=10, face = "italic",
colour = "black"),
legend.position = "none")
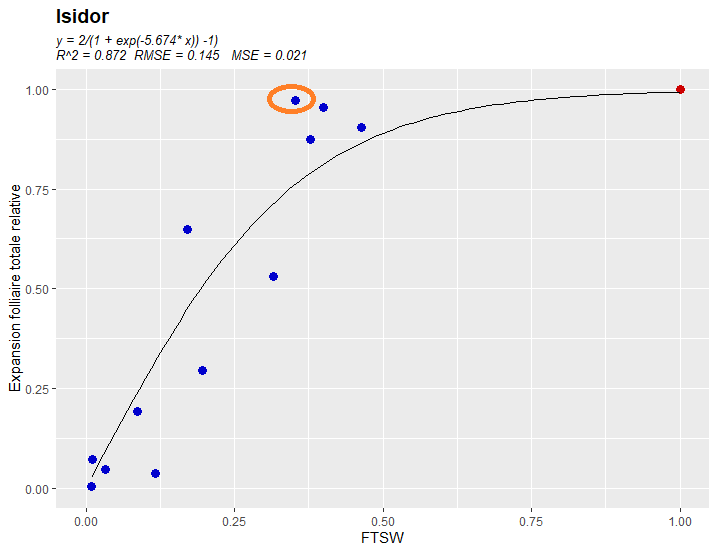
Now I want to calculate RMSE and standard deviation between the coordinate point circled by orange (0.352560790, 0.970762905) and the non-linear regression y = 2/(1 exp(-5.674* x))-1 (the coordinate point in the regression line with the same abscissa 0.352560790). Could anyone please give me suggestions?
Thank you so much!
CodePudding user response:
You didn't show us how you made your model, so I'm assuming it's something like this:
mod <- nls(NLE ~ 2/(1 exp(a * FTSW_apres_arros)) - 1, start = list(a = -6),
data = ISIDOR)
We can use our model to insert the expected value of NLE for each row in our original data frame like this:
ISIDOR$pred <- predict(mod)
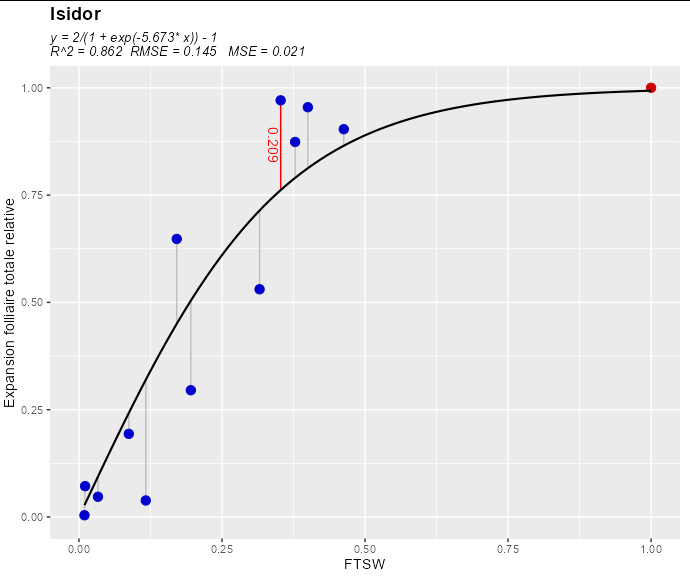
And we can get the residuals (or error) of each point (i.e. its distance from the regression line like this:
ISIDOR$error <- summary(mod)$residuals
This results in the following data frame:
head(ISIDOR)
#> # A tibble: 6 x 7
#> Pos_heliaphen traitement Variete FTSW_apres_arros NLE pred error
#> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 W30 WW Isidor 0.463 0.903 0.865 0.0385
#> 2 X41 WW Isidor 0.400 0.955 0.813 0.142
#> 3 Y27 WW Isidor 0.353 0.971 0.762 0.209
#> 4 Z24 WW Isidor 0.378 0.874 0.790 0.0837
#> 5 Y27 WW Isidor 0.171 0.648 0.450 0.198
#> 6 W30 WW Isidor 0.316 0.531 0.714 -0.184
For our plot, we calculate the various metrics and use geom_smooth to plot our regression line (this means we don't need a prediction data frame)
library(geomtextpath)
Rsquared <- 1 - var(ISIDOR$error) / var(ISIDOR$NLE)
MSE <- mean(ISIDOR$error^2)
RMSE <- sqrt(MSE)
ggplot(ISIDOR, aes(FTSW_apres_arros, NLE))
geom_segment(aes(xend = FTSW_apres_arros, yend = pred), color = 'gray')
geom_textsegment(aes(xend = FTSW_apres_arros, yend = pred,
label = round(error, 3)),
color = 'red', data = ISIDOR[3,], vjust = 1.1)
geom_point(aes(color = Variete), shape = 19, cex = 3)
geom_smooth(method = nls, formula = y ~ 2/(1 exp(a * x)) - 1,
method.args = list(start = list(a = -6)), se = FALSE,
color = 'black', size = 0.75)
scale_color_manual(values = c("red3", "blue3"))
scale_y_continuous(limits = c(0, 1.0))
scale_x_continuous(limits = c(0, 1))
labs(title = "Isidor",
y = "Expansion folliaire totale relative",
x = "FTSW",
subtitle = paste0("y = 2/(1 exp(", round(coef(mod), 3), "* x)) - 1",
"\n", "R^2 = ", round(Rsquared, 3)," RMSE = ",
round(RMSE, 3), " MSE = ", round(MSE, 3)))
theme(plot.title = element_text(hjust = 0, size = 14, face = "bold",
colour = "black"),
plot.subtitle = element_text(hjust = 0,size=10, face = "italic",
colour = "black"),
legend.position = "none")