Within a Shiny application, I would like to provide users the option to select a regression method and then show the fit on their data on an existing ggplot2 graph (showing the original data as a geom_step) --> so the graph already exists!
There is a restriction on this graph as colours and line types are mapped to two crossed variables. Yet the combination between the variables ought to be shown as 1 legend item.
This is the dummy data:
library(tidyverse)
createGroup <- function(group, category, effect){
x <- seq(1,10)
data.frame(
time = x,
y = effect * x rnorm(10, mean = 0, sd = 0.1),
group = group,
cat = category
)
}
set.seed(12)
ipt <- list(group = paste0('Arm ', rep(c(1,2), each = 2)),
category = rep(LETTERS[1:2], 2),
effect = c(0.04, 0.09, 0.35, 0.45))
tmp <- lapply(1:4, \(x) do.call(createGroup,
list(group = ipt$group[x], category = ipt$category[x], effect = ipt$effect[x])))
DF <- do.call('rbind', tmp)
DF$group <- factor(DF$group)
# To combine legends, pasting the group and category so that colour and line type
# can be mapped
DF$combined <- paste0(DF$group, ' & cat ', DF$cat)
DF
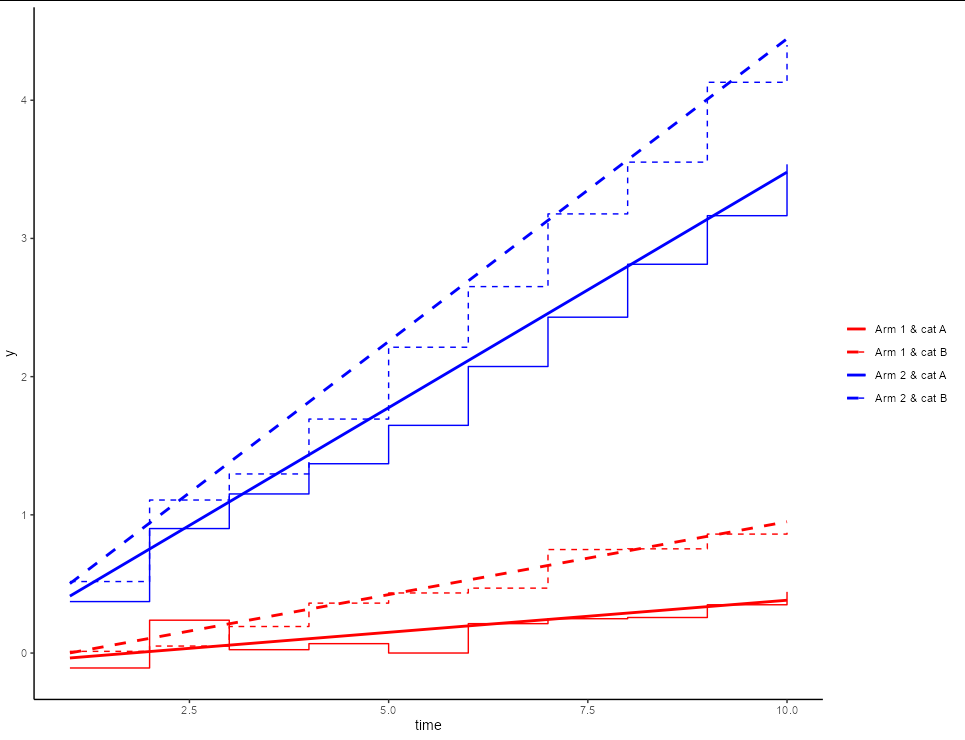
Producing this graph:
p1 <- ggplot(DF, aes(x = time, y = y))
geom_step(aes(colour = combined, linetype = combined))
scale_colour_manual('', values = c('red', 'red', 'blue', 'blue'),
breaks = unique(DF$combined))
scale_linetype_manual('', values = c(1,2,1,2),
breaks = unique(DF$combined))
theme_classic()
p1
Now fitting a linear regression in each combined group:
getPred <- function(x, id, method){
fit <- lm(y ~ time, data = data.frame(x))
data.frame(y = predict(fit, newdata = data.frame(time = 1:10)), time = 1:10, method = paste(method, id))
}
# Create a DF with the predictions, limited to current time range.
preds <- DF %>%
group_by(combined) %>%
tidyr::nest() %>%
mutate(pred = purrr::map(data, ~getPred(x = ., id = combined, method = 'lm')))
predDF <- do.call('rbind', preds$pred)
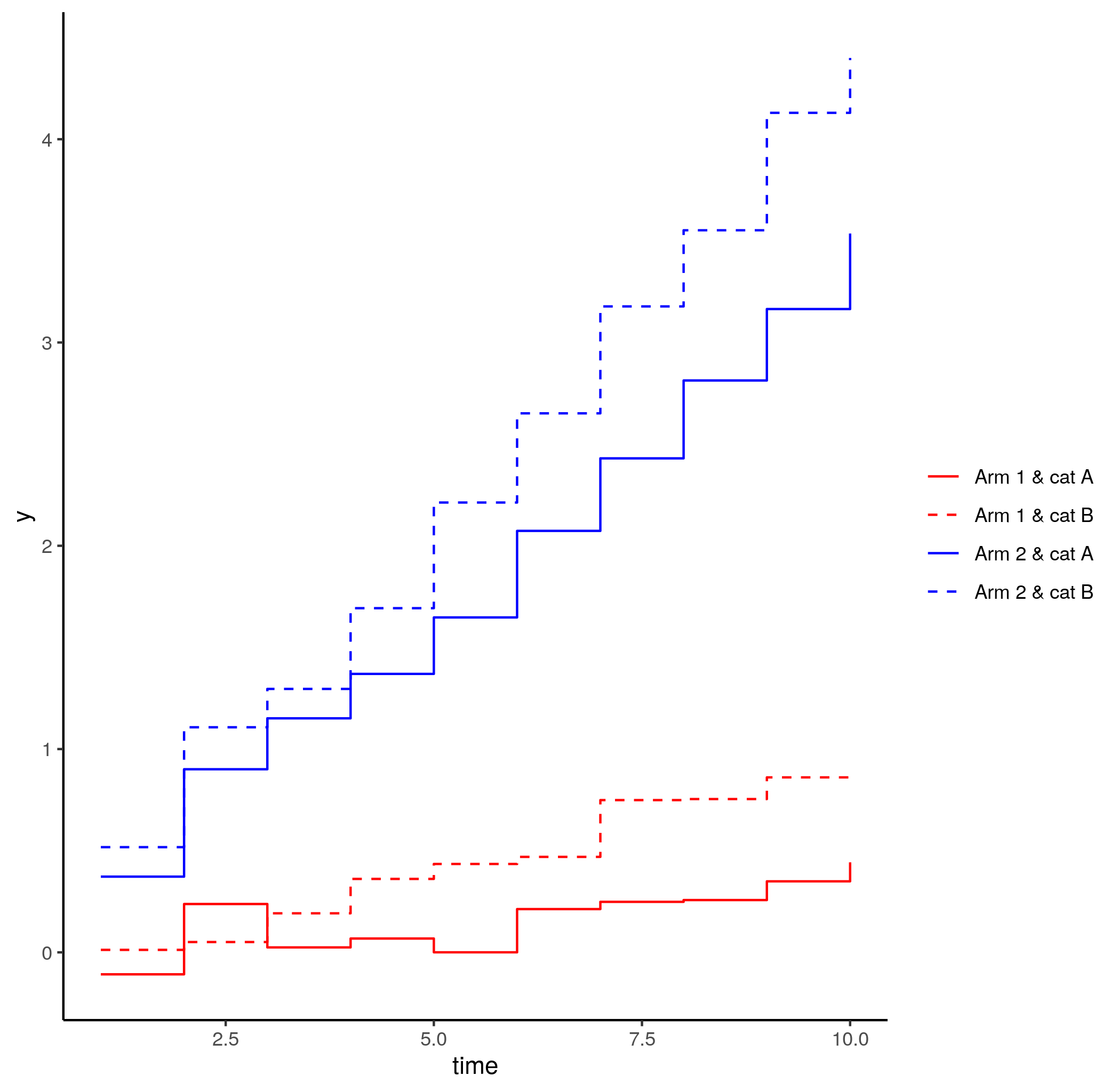
One can plot the new lines by method:
p1 geom_line(data = predDF, aes(x = time, y = y, group = method))
This gives you this:
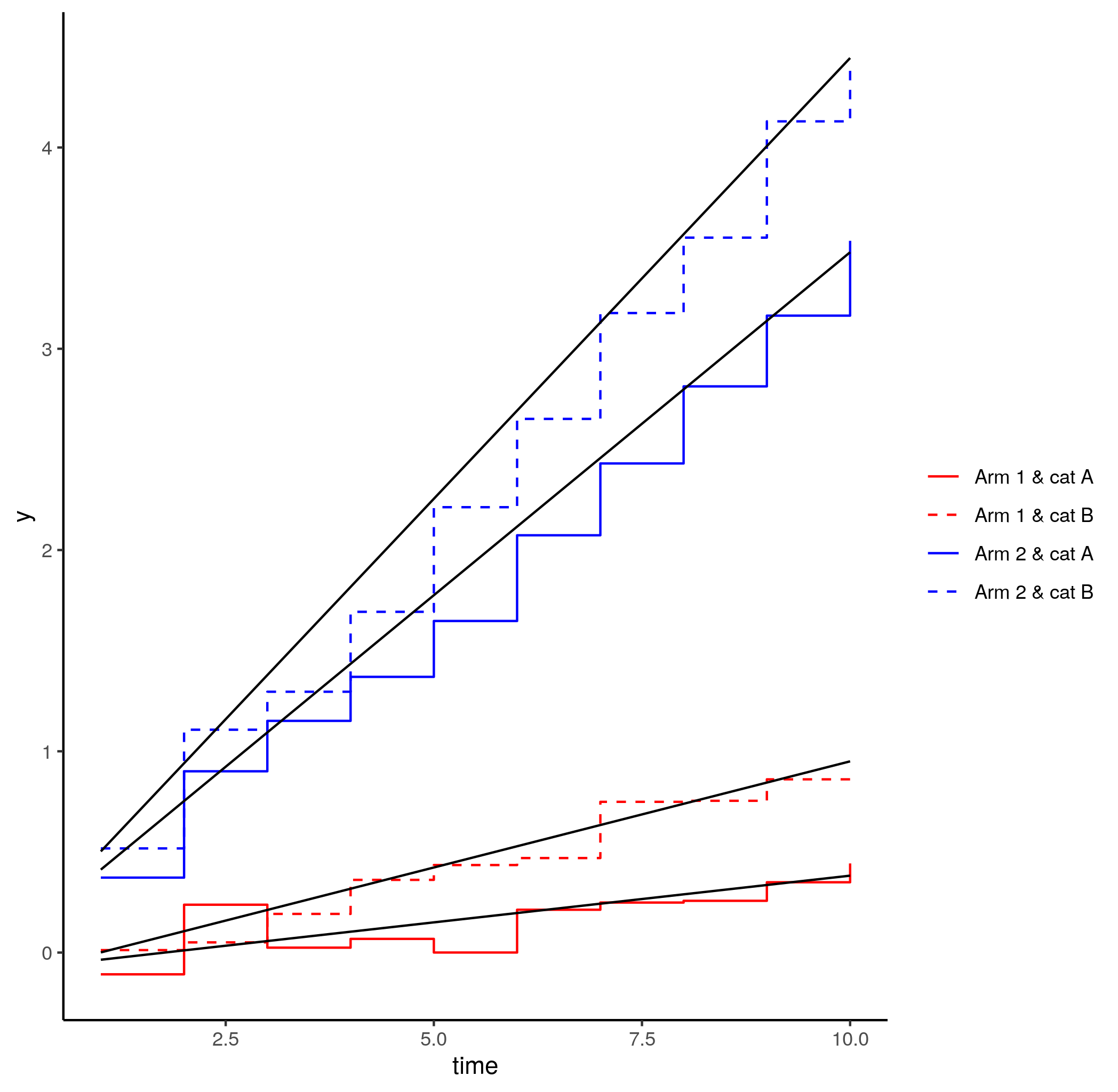
However, the original plot does not have the colours (nor line types) mapped to the (new) levels of the method column (as they did not exist). Hence re-assigning colour for the predDF to 'method' does not work.
p1 geom_line(data = predDF, aes(x = time, y = y, colour = method))
Error: Insufficient values in manual scale. 8 needed but only 4 provided.
Hence: is there any way to strip the mapping from a ggplot2 object and re-do it in a later stage? Or is there no other option than to re-build the entire graph (after binding the DF with predDF)?
CodePudding user response:
You're reinventing the wheel a hit here. You don't need to create a prediction data frame. Instead, use method = lm inside a geom_smooth call. For colors and line types, you can borrow the aesthetic mapping from the first layer of p1:
p1 geom_smooth(mapping = p1$layers[[1]]$mapping, formula = y~ x, method = lm,
se = FALSE)
CodePudding user response:
Next to the answer from Allan, I also want to provide this simple solution I found the other day:
p1 scale_colour_manual('', values = rep(c('red', 'red', 'blue', 'blue'),2),
breaks = c(unique(DF$combined), unique(predDF$method)))
scale_linetype_manual('', values = rep(c(1,2,1,2),2),
breaks = c(unique(DF$combined), unique(predDF$method)))
geom_line(data = predDF, aes(x = time, y = y, colour = method, linetype = method))
It is only a matter of doing the re-mapping first, then adding new lines...