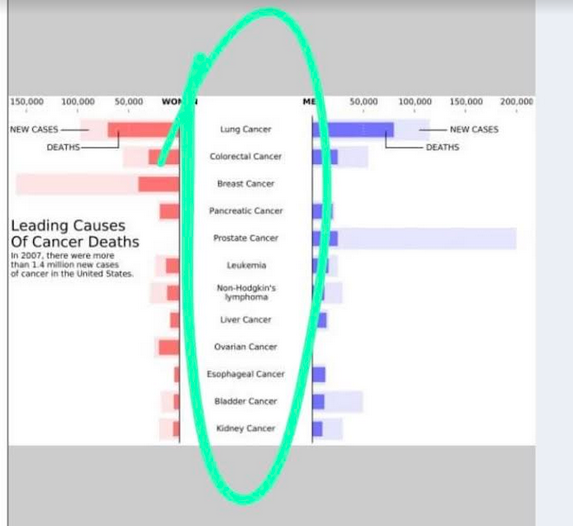
I am trying to replicate a fantastic plot I found in a paper. The plot basically has a central y axis and then bars according to a variable are created in both left and right side. The plot is next:
I have a similar dataframe to replicate this plot. My dataframe df has next structure:
df
# A tibble: 64 x 4
Gender Cases Deaths Group
<chr> <int> <dbl> <chr>
1 F 6163 24 G1
2 M 9067 136 G1
3 F 430 4 G2
4 M 1026 51 G2
5 F 43 0 G3
6 M 67 1 G3
7 F 1382 43 G4
8 M 888 26 G4
9 F 249 4 G5
10 M 191 10 G5
# ... with 54 more rows
I would like to create the mentioned plot, showing in the central axis the variable Group and in the x axis (left and right) I would like to show the variables Cases and Deaths according to Gender variable, this would be bars to left for M gender and bars to right for F gender.
In the attempt to reach the objective plot I sketched some code that could be the base but I do not know how to modify it in order to change the order of axis. This is the code:
library(ggplot2)
library(tidyverse)
#Code for F gender
df %>% pivot_longer(-c(Group,Gender)) %>%
filter(Gender=='F') %>%
mutate(Group=factor(Group,levels = unique(df$Group),ordered = T)) %>%
ggplot(aes(x=Group,y=value,fill=name))
geom_bar(stat = 'identity')
scale_x_discrete(limits = rev(unique(df$Group)))
coord_flip()
theme(legend.position = 'top')
#Code for F gender
df %>% pivot_longer(-c(Group,Gender)) %>%
filter(Gender=='M') %>%
mutate(Group=factor(Group,levels = unique(df$Group),ordered = T)) %>%
ggplot(aes(x=Group,y=value,fill=name))
geom_bar(stat = 'identity')
scale_x_discrete(limits = rev(unique(df$Group)))
coord_flip()
theme(legend.position = 'top')
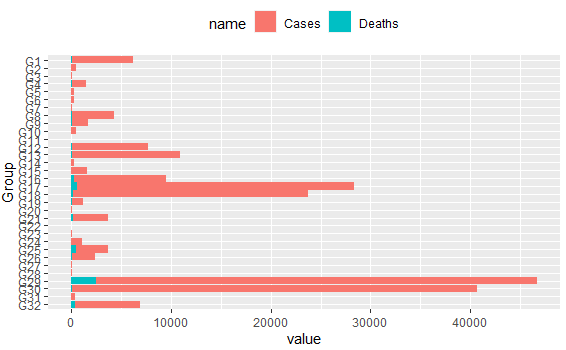
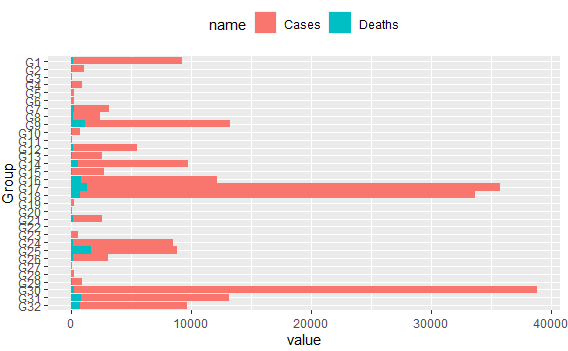
Which produces next plots:
An issue I found with these plots is that despite showing the bars as I want, they have two problems. First, I would like that one of the bars (ex. Deaths) could be thinner than the other bar (ex. Cases). Second, the structure of the bars is accumulating all values. I would like that both measures start from zero in each axis.
I hope this plot can be created in ggplot2. My data df is next:
#Data
df <- structure(list(Gender = c("F", "M", "F", "M", "F", "M", "F",
"M", "F", "M", "F", "M", "F", "M", "F", "M", "F", "M", "F", "M",
"F", "M", "F", "M", "F", "M", "F", "M", "F", "M", "F", "M", "F",
"M", "F", "M", "F", "M", "F", "M", "F", "M", "F", "M", "F", "M",
"F", "M", "F", "M", "F", "M", "F", "M", "F", "M", "F", "M", "F",
"M", "F", "M", "F", "M"), Cases = c(6163L, 9067L, 430L, 1026L,
43L, 67L, 1382L, 888L, 249L, 191L, 278L, 248L, 36L, 2925L, 4248L,
2286L, 1576L, 12106L, 441L, 690L, 7L, 53L, 7645L, 5335L, 10862L,
2546L, 229L, 9136L, 1578L, 2657L, 9301L, 11384L, 27773L, 34435L,
23599L, 32952L, 1105L, 170L, 31L, 94L, 3469L, 2408L, 1L, 6L,
86L, 566L, 1108L, 8355L, 3203L, 7174L, 2314L, 2943L, 46L, 54L,
26L, 187L, 44201L, 837L, 40608L, 38616L, 343L, 12284L, 6571L,
8882L), Deaths = c(24, 136, 4, 51, 0, 1, 43, 26, 4, 10, 0, 2,
1, 242, 84, 112, 49, 1164, 7, 33, 0, 4, 26, 115, 63, 24, 7, 556,
14, 86, 228, 784, 596, 1344, 189, 705, 24, 15, 0, 1, 180, 120,
0, 0, 0, 7, 8, 155, 465, 1630, 39, 125, 3, 3, 0, 0, 2511, 87,
114, 219, 8, 847, 340, 760), Group = c("G1", "G1", "G2", "G2",
"G3", "G3", "G4", "G4", "G5", "G5", "G6", "G6", "G7", "G7", "G8",
"G8", "G9", "G9", "G10", "G10", "G11", "G11", "G12", "G12", "G13",
"G13", "G14", "G14", "G15", "G15", "G16", "G16", "G17", "G17",
"G18", "G18", "G19", "G19", "G20", "G20", "G21", "G21", "G22",
"G22", "G23", "G23", "G24", "G24", "G25", "G25", "G26", "G26",
"G27", "G27", "G28", "G28", "G29", "G29", "G30", "G30", "G31",
"G31", "G32", "G32")), row.names = c(NA, -64L), class = c("tbl_df",
"tbl", "data.frame"))
Many thanks!
CodePudding user response:
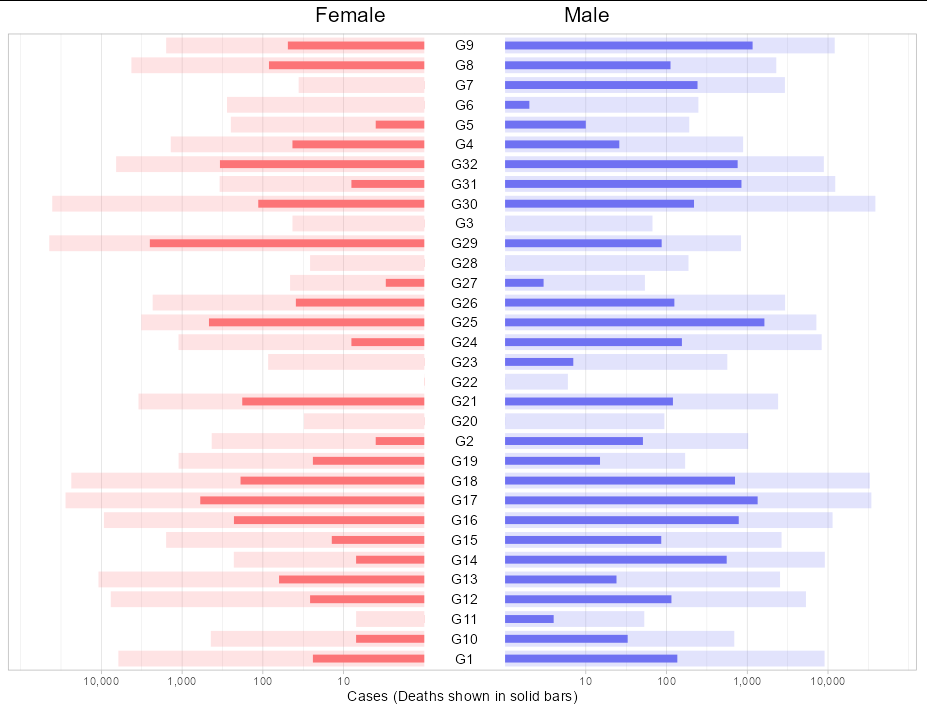
I wouldn't pivot here at all. If you keep cases and deaths separate, two calls to geom_col will overlap. To get the mirror image setup with a central y axis, I would simply draw a rectangle and the labels as annotations. This means you need to fake your x axis too, to adjust for the space the rectangle takes up.
You will need to make the x values for one of the genders negative. Since we need a log transform, make them negative after taking logs.
df %>%
mutate(Cases = ifelse(Gender == "F", -log10(Cases) - 0.5, log10(Cases) 0.5),
Deaths = ifelse(Deaths == 0, 1, Deaths),
Deaths = ifelse(Gender == "F", -log10(Deaths) - 0.5,
log10(Deaths) 0.5)) %>%
ggplot(aes(Cases, Group, fill = Gender))
geom_col(width = 0.8, alpha = 0.2)
geom_col(width = 0.4, aes(x = Deaths))
annotate(geom = "rect", xmin = -0.5, xmax = 0.5, ymax = Inf, ymin = -Inf,
fill = "white")
annotate(geom = "text", x = 0, y = levels(factor(df$Group)),
label = levels(factor(df$Group)))
theme_light()
scale_x_continuous(breaks = c(-4.5, -3.5, -2.5, -1.5, 1.5, 2.5, 3.5, 4.5),
labels = c("10,000", "1,000", "100", "10", "10",
"100", "1,000", "10,000"))
scale_fill_manual(values = c("#fc7477", "#6f71f2"))
labs(title = "Female Male",
x = "Cases (Deaths shown in solid bars)")
theme(axis.text.y = element_blank(),
axis.title.y = element_blank(),
axis.ticks.y = element_blank(),
panel.grid.major.y = element_blank(),
plot.title = element_text(size = 16, hjust = 0.5),
legend.position = "none")