I am attempting to run a model using glmmTMB. When I include avgt60, it does weird things in the model and I am not really sure why. When I include it as a non poly term, it gives me NaN values. When I include it as a poly() term, it throws the entire model off. When I exclude it, it seems to be fine... I am new to this type of work so any advice is appreciated!
m1 <- glmmTMB(dsi ~ poly(rh60, degree = 2) poly(wndspd60, degree = 2) poly(raintt60, degree = 2) avgt60 (1|year) (1|site),
family = "nbinom2", data = weather1)
I get:
Family: nbinom2 ( log )
Formula: dsi ~ poly(rh60, degree = 2) poly(wndspd60, degree = 2) poly(raintt60, degree = 2) avgt60 (1 | year) (1 | site)
Data: weather1
AIC BIC logLik deviance df.resid
1647.9 1687.9 -813.0 1625.9 269
Random effects:
Conditional model:
Groups Name Variance Std.Dev.
year (Intercept) 5.883e-24 2.426e-12
site (Intercept) 6.396e-07 7.997e-04
Number of obs: 280, groups: year, 3; site, 6
Dispersion parameter for nbinom2 family (): 0.232
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -7.8560 NaN NaN NaN
poly(rh60, degree = 2)1 47.9631 NaN NaN NaN
poly(rh60, degree = 2)2 -5.4370 NaN NaN NaN
poly(wndspd60, degree = 2)1 61.7092 NaN NaN NaN
poly(wndspd60, degree = 2)2 -74.9432 NaN NaN NaN
poly(raintt60, degree = 2)1 27.2669 NaN NaN NaN
poly(raintt60, degree = 2)2 -72.9072 NaN NaN NaN
avgt60 0.4384 NaN NaN NaN
But, without the avgt60 variable...
m1 <- glmmTMB(dsi ~ poly(rh60, degree = 2) poly(wndspd60, degree = 2) poly(raintt60, degree = 2) (1|year) (1|site),
family = "nbinom2", data = weather1)
Family: nbinom2 ( log )
Formula: dsi ~ poly(rh60, degree = 2) poly(wndspd60, degree = 2) poly(raintt60, degree = 2) (1 | year) (1 | site)
Data: weather1
AIC BIC logLik deviance df.resid
1648.2 1684.6 -814.1 1628.2 270
Random effects:
Conditional model:
Groups Name Variance Std.Dev.
year (Intercept) 2.052e-10 1.433e-05
site (Intercept) 4.007e-10 2.002e-05
Number of obs: 280, groups: year, 3; site, 6
Dispersion parameter for nbinom2 family (): 0.23
Conditional model:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.3677 0.3482 3.928 8.56e-05 ***
poly(rh60, degree = 2)1 23.8058 9.6832 2.458 0.013953 *
poly(rh60, degree = 2)2 -0.3452 4.2197 -0.082 0.934810
poly(wndspd60, degree = 2)1 34.4332 10.1328 3.398 0.000678 ***
poly(wndspd60, degree = 2)2 -61.2044 6.5179 -9.390 < 2e-16 ***
poly(raintt60, degree = 2)1 12.0109 6.4949 1.849 0.064417 .
poly(raintt60, degree = 2)2 -57.2197 6.0502 -9.457 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
If I leave avgt60 in as a poly() term, it throws the entire model off, and nothing is significant. Any thoughts here?
Here's a link to the dataset, with site names redacted: 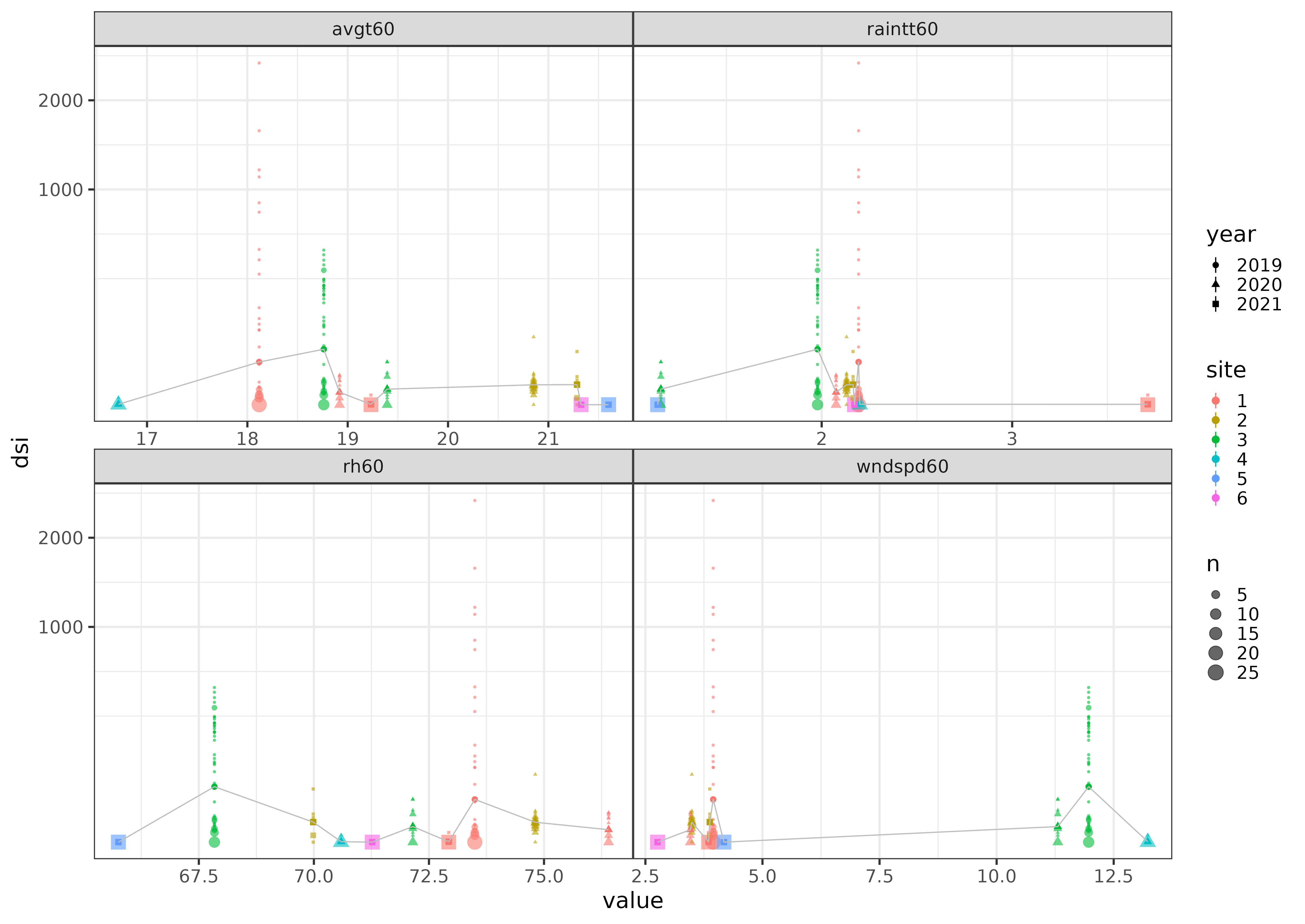
dsivalues for sites 5 and 6 are always zero (only measured in 2021)dsivalues are very high in 2019, only two sites (1 and 3) measured- there is no particular pattern, and certainly not one that's not confounded with site and year.
I would probably try to draw only qualitative conclusions, or very simple quantitative conclusions, from these data ...
library(tidyverse); theme_set(theme_bw())
w3 <- (weather1
|> as_tibble()
|> select(-date)
|> pivot_longer(-c(site, year, dsi), names_to = "var")
|> mutate(across(c(year,site), factor))
)
theme_set(theme_bw(base_size = 20) theme(panel.spacing = grid::unit(0, "lines")))
(ggplot(w3)
aes(x = value, y = dsi, colour = site, shape = year)
stat_sum(alpha = 0.6)
stat_summary(fun = mean)
stat_summary(fun = mean, geom = "line", aes(group = 1), colour = "gray")
facet_wrap(~var, scale = "free_x")
scale_y_sqrt()
)
Murtaugh, Paul A. “Simplicity and Complexity in Ecological Data Analysis.” Ecology 88, no. 1 (2007): 56–62.
