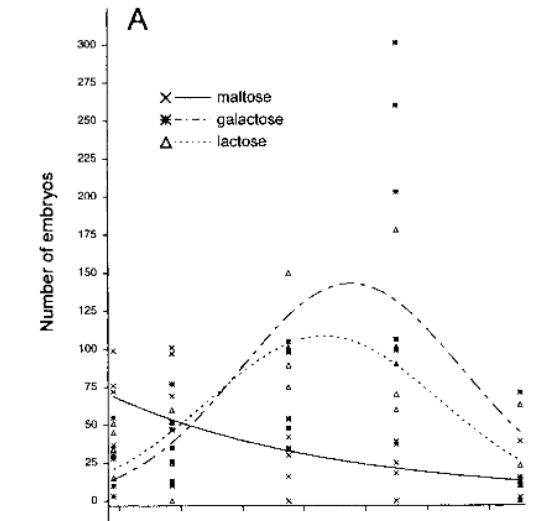
I'm trying to plot the observed vs. the estimated line of a negative binomial regression model with random effects, where this curve takes into account the structure of the adopted model.
The data and the model were fitted as follows:
d.AD <- data.frame(treatment = gl(12,12),
treatment2 = gl(4,1,36),
counts = rpois(144, 4.03),
treatment3 = gl(3,4,36),
ID = gl(12,12))
d.AD$treatment2 = as.integer(d.AD$treatment2)
d.AD$treatment3 = as.factor(d.AD$treatment3)
d.AD$ID = as.factor(d.AD$ID)
library(ggplot2)
library(lme4)
mode1A <- glmer.nb(counts~treatment3*poly(treatment2, 2) (1|ID),
data=d.AD, verbose=F)
The graph I intend to perform is something like this:
The attempt was as follows:
d.AD$de6 <- with(d.AD,counts)
ggplot(d.AD, aes(treatment3, treatment2, de6))
geom_point()
geom_point(aes(y = fitted(mode1A)), colour = "black")
geom_line(aes(y = predict(mode1A, d.AD["treatment3"], d.AD["treatment2"],
type = "response")), colour = "black")
theme(axis.text.x = element_text(size = 20,color = "black"),
axis.text.y = element_text(size = 20,color = "black"),
axis.text = element_text(size = 25))
However, I am facing the following error:
Error in setParams(object, newparams) : params should be specifed as a list with elements from {"beta", "theta"}
CodePudding user response:
How about this:
d.AD <- data.frame(treatment = gl(12,12),
treatment2 = gl(4,1,36),
counts = rpois(144, 4.03),
treatment3 = gl(3,4,36),
ID = gl(12,12))
d.AD$treatment2 = as.integer(d.AD$treatment2)
d.AD$treatment3 = as.factor(d.AD$treatment3)
d.AD$ID = as.factor(d.AD$ID)
library(ggplot2)
library(lme4)
#> Loading required package: Matrix
library(ggeffects)
mode1A <- glmer.nb(counts~treatment3*poly(treatment2, 2) (1|ID),
data=d.AD, verbose=F)
#> Warning in theta.ml(Y, mu, weights = object@resp$weights, limit = limit, :
#> iteration limit reached
e <- ggpredict(mode1A, terms=c("treatment2 [all]", "treatment3"), type="re")
plot(e, rawdata=TRUE, ci=FALSE)
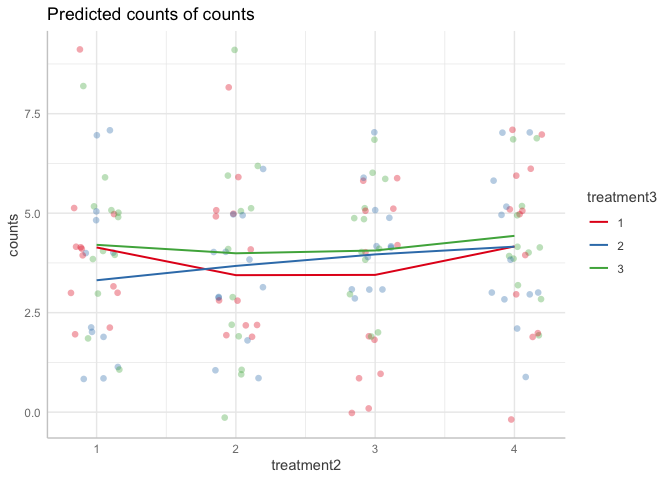
Created on 2022-06-05 by the reprex package (v2.0.1)