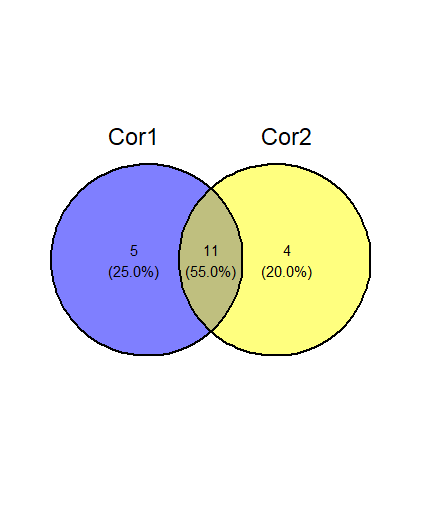
I have a DataFrame df1 and I want to make a pairwise Venn diagram.
I want to show the number of overlap of the ENSGs (which should be defined by the treshold Cor > 0.1) from Cor1 and Cor2.
df1 <- structure(list(ENSG = c("ENSG89306", "ENSG230107",
"ENSG128274", "ENSG100266", "ENSG100300", "ENSG100290",
"ENSG100347", "ENSG261251", "ENSG138964", "ENSG93000",
"ENSG226328", "ENSG100376", "ENSG205643", "ENSG128408",
"ENSG77935", "ENSG130638", "ENSG241990", "ENSG75234",
"ENSG260708", "ENSG54611", "ENSG0128159", "ENSG188511",
"ENSG212939", "ENSG100258", "ENSG235111", "ENSG184164",
"ENSG79974", "ENSG170638", "ENSG73169", "ENSG100427"), Cor1 = c(0.143714860528127, 0.140903769519937,
0.220285794778746, -0.0764275174733086, 0.189080738386014, 0.303454798874576,
0.268021621489439, 0.160662468776058, 0.0797970266014118, 0.0303382173551934,
0.0113128251324056, 0.139860538051072, 0.018553280666828, 0.219125961518781,
0.0898644147097944, 0.0595462431532415, 0.0161782451465834, 0.0633049687607816,
0.0774210516121433, 0.548879883295163, 0.133707267647068, -0.0275291634151178,
0.0132847938530041, -0.0468134127145183, 0.212056295268778, 0.169525452308467,
0.223304209279054, 0.0554950703076702, 0.268747518524584, 0.210788129456603),
Cor2 = c(-0.0567563530541253, 0.1467342043789, -0.00462978967736684, -0.0949858201084905, -0.0128046412985325, 0.308707458359154,
-0.0288326568108287, 0.123139258225873, 0.127834335556582, 0.0476613529381162,
0.108169226237846, 0.135726417808339, -0.0499017880297932, 0.269937039031533,
0.124331777264371, -0.00443812208979526, 0.0531319792200424,
0.0639288256005855, 0.188225249289256, 0.471541813383384, 0.116236004671794,
0.0197695247788121, 0.0476613529381162, -0.0501695121118335,
0.194805149886956, 0.148499060471642, 0.263555546219041, 0.0374697142901511,
0.213968479801099, 0.00159814317995886)), row.names = c(NA, 30L), class = "data.frame")
I do appreciate your help.
CodePudding user response:
Perhaps you mean to do something like this, leveraging the ggvenn package?. mutate(across()) here converts Cor1 and Cor2 columns into logical columns; these can be directly fed to ggvenn, and the overlap region is the number of rows where all (in this case "both") logical vectors are TRUE.
library(dplyr); library(ggvenn)
df1 %>%
mutate(across(Cor1:Cor2, ~(.x>0.1))) %>%
ggvenn(c("Cor1", "Cor2"))