I'm trying to make multiple caterpillar plots for different dependent variables but same independent variables by ggplot.
With this trial data,
x1 <- c(1,2,3,4,5)
x2 <- c(4,5,6,1,2)
x3 <- c(7,8,3,2,1)
x4 <- c(3,4,5,6,7)
y1 <- c(10,9,11,18,13)
y2 <- c(12,20,21,19,11)
y3 <- c(18,16,12,11,18)
df <- base::data.frame(x1, x2, x3, x4, y1, y2, y3)
at first I tried to make the plots without facet .
lm1 <- lm(data = df,
y1 ~ x1 x2 x3 x4)
lm_plot1 <- lm %>%
tidytext::tidy() %>%
ggplot2::ggplot(aes(x = term, y = estimate))
# Make two more lm_plots for y2, y3 in the same way...
# And put them on one screen by grid.arrange()
gridExtra::grid.arrange(lm_plot1, lm_plot2, lm_plot3)
Now I'd like to make faceted image. So I tried to make longer-data
df %>%
tidyr::pivot_longer(c(y1,y2,y3)) %>%
dplyr::rename(dv = name) %>%
group_by(dv)
but I don't know how to estimate each model after group_by function and to make faceted plots by ggplot.
CodePudding user response:
One approach is to nest() the data using tidyr.
First let's load libraries and make some better example data:
library(dplyr)
library(tidyr)
library(ggplot2)
library(purrr)
set.seed(1001)
df1 <- matrix(sample(1:100, 350, replace = TRUE),
ncol = 7,
dimnames = list(NULL, c("x1", "x2", "x3", "x4", "y1", "y2", "y3")))
df1 <- as.data.frame(df1)
If we pivot_longer and nest on name, we get nested tibbles for each of the y-values. They contain a value column with the value for y1, y2, y3.
df1 %>%
pivot_longer(cols = starts_with("y")) %>%
nest(data = -name)
name data
<chr> <list>
1 y1 <tibble [50 × 5]>
2 y2 <tibble [50 × 5]>
3 y3 <tibble [50 × 5]>
Now we can use purrr::map to make a model for each y-value:
df1 %>%
pivot_longer(cols = starts_with("y")) %>%
nest(data = -name) %>%
mutate(model = map(data, ~lm(value ~ x1 x2 x3 x4, data = .)),
tidydata = map(model, tidy))
name data model tidydata
<chr> <list> <list> <list>
1 y1 <tibble [50 × 5]> <lm> <tibble [5 × 5]>
2 y2 <tibble [50 × 5]> <lm> <tibble [5 × 5]>
3 y3 <tibble [50 × 5]> <lm> <tibble [5 × 5]>
Finally, remove unwanted columns and unnest the nested tidied output:
df1 %>%
pivot_longer(cols = starts_with("y")) %>%
nest(data = -name) %>%
mutate(model = map(data, ~lm(value ~ x1 x2 x3 x4, data = .)),
tidydata = map(model, tidy)) %>%
select(-data, -model) %>%
unnest(cols = "tidydata")
# A tibble: 15 × 6
name term estimate std.error statistic p.value
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 y1 (Intercept) 41.1 12.9 3.17 0.00272
2 y1 x1 -0.0379 0.134 -0.283 0.778
3 y1 x2 0.0889 0.142 0.628 0.533
4 y1 x3 0.0979 0.155 0.632 0.530
5 y1 x4 0.144 0.151 0.954 0.345
6 y2 (Intercept) 69.7 13.8 5.04 0.00000810
7 y2 x1 0.00506 0.143 0.0354 0.972
8 y2 x2 -0.251 0.151 -1.66 0.105
9 y2 x3 0.00999 0.166 0.0603 0.952
10 y2 x4 -0.278 0.161 -1.73 0.0910
11 y3 (Intercept) 54.7 14.6 3.75 0.000503
12 y3 x1 0.0645 0.150 0.428 0.670
13 y3 x2 -0.0406 0.159 -0.255 0.800
14 y3 x3 -0.0645 0.174 -0.370 0.713
15 y3 x4 -0.0788 0.170 -0.465 0.644
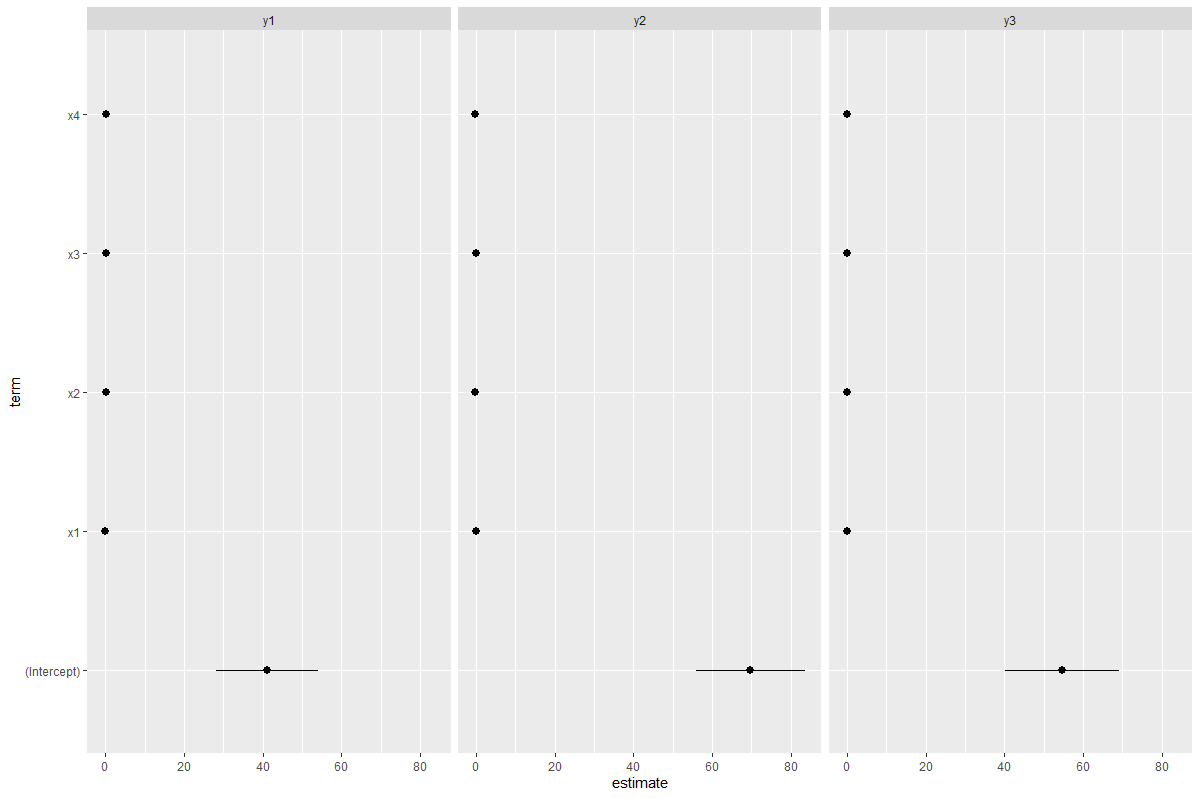
Now we can ggplot with facets. I'm assuming that your "caterpillar plot" is similar to a "forest plot". So something like this:
df1 %>%
pivot_longer(cols = starts_with("y")) %>%
nest(data = -name) %>%
mutate(model = map(data, ~lm(value ~ x1 x2 x3 x4, data = .)),
tidydata = map(model, tidy)) %>%
select(-data, -model) %>%
unnest(cols = "tidydata") %>%
ggplot(aes(term, estimate))
geom_point()
geom_pointrange(aes(ymin = estimate - std.error,
ymax = estimate std.error))
coord_flip()
facet_wrap(~name)
Result: