i have a little problem with my code and i can not see it.
I am trying to do a linear regression with some data. Until now, i am using np.polyfit()and sklearn LinearRegression(). Both regressions work but when i do the plotting, i can see that the intercept coefficient does not match what the plot shows and honestly i don´t why.
So, here is my code:
# Using np.polyfit()
x_to_plot = np.array([3.1685206 , 2.25510996, 2.96102541, 2.88536998, 3.19026643,
3.35665069, 3.62661427, 3.19285197, 2.57931189, 3.14756505,
3.99712702, 4.08227484, 3.14058442, 4.4395179 , 3.8499418 ,
3.812325 , 3.9126594 , 3.85465249, 4.57109736, 4.25932984,
4.07329771, 3.48126781, 3.39053495, 4.00443561, 3.38871004,
3.88689312, 4.39439235, 3.73192345, 3.07600559, 4.02592055,
3.66448407, 3.62788865, 3.12075542, 3.67316965, 3.59271334,
4.22789875, 3.7891345 , 4.36675152, 3.45593254, 2.6429717 ,
3.25813122])
y_to_plot = np.array([2.2 , 2.27 , 2.4 , 2.36 , 2.44 ,
2.67 , 3.11 , 3.11 , 2.19 , 2.77 ,
3.3 , 3.14 , 2.63 , 2.59549622, 3.05728564,
2.73239376, 3.12319808, 2.72835378, 3.4476231 , 3.1693805 ,
3.16375752, 2.84571802, 2.89486966, 2.96801571, 2.90254678,
3.03100428, 3.14612804, 2.97726621, 2.8785218 , 3.20628604,
2.50650503, 2.53275438, 2.59106461, 3.00043408, 2.76715587,
3.01283722, 2.36172784, 3.31868927, 2.65896484, 2.0211893 ,
2.93399316])
fit = np.polyfit(x_to_plot, y_to_plot, 1)
scolpe = fit[0]
intercept = fit[1]
fit_eq = scolpe* x_to_plot intercept
# plot
plt.figure(1)
plt.xlabel("X")
plt.ylabel("Y")
plt.plot(x_to_plot, y_to_plot, marker="o")
plt.plot(x_to_plot, fit_eq, "--k")
plt.show()
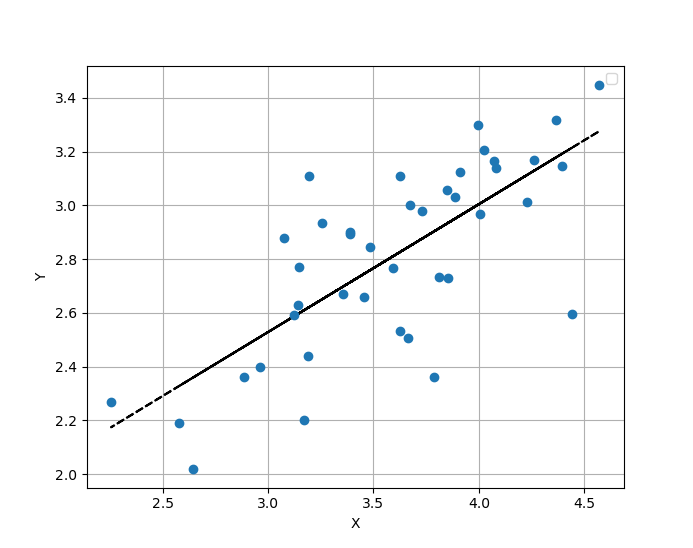
Then, if i print the intercept value, it says that intercept = 1.102355...
But you can see in the plot that intercept value has to be $\approx$ 2.2.
I also used plt.ylim() but it did´nt work.
I don't know if this detail is important (i don't think so, but here is it): x_to_plot and y_to_plot are like scale arrays)? i mean, previously i had the data for x-y axis, then i applied np.log10() and then i obtained x_to_plot and y_to_plot.
I don´t add the code for the sklearn regression, because i feel its the same thing.
CodePudding user response:
Your y axis needs to be at x = 0 for the intercept to match your model output.
i.e. the intercept of a linear model is the output when all predictors equal 0. It doesn’t always (or even usually) make sense for graphs to default so that the y axis occurs at x = 0, as that might be outside the range of the data (as in this case).
If you force the x axis to include 0, and allow the y axis to occur at x = 0, then the y intercept would indeed match the model output.