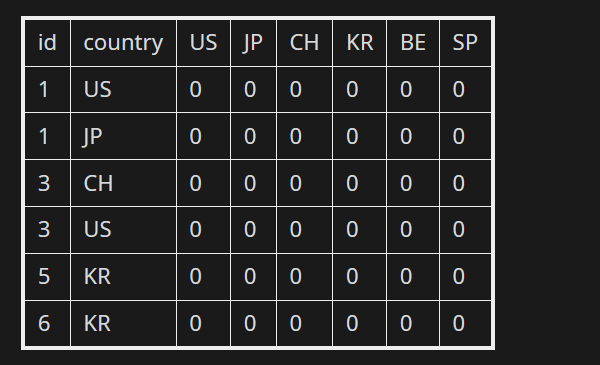
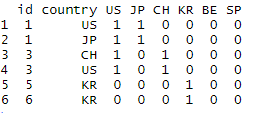
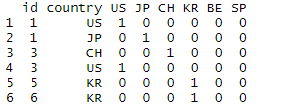
This is how my data looks:
My dput is given below as:
structure(list(id = c(1, 1, 3, 3, 5, 6), country = c("US", "JP","CH", "US", "KR", "KR"), US = c(0, 0, 0, 0, 0, 0), JP = c(0,0, 0, 0, 0, 0), CH = c(0, 0, 0, 0, 0, 0), KR = c(0, 0, 0, 0,0, 0), BE = c(0, 0, 0, 0, 0, 0), SP = c(0, 0, 0, 0, 0, 0)), class = "data.frame", row.names = c(NA, -6L))
This is what I want:
Running data2[levels(factor(data2$country))] = model.matrix(~country - 1, data2) gives me a close but WRONG result:
How can I get the structure that I'm looking for? Please help; I've been working on this problem for days and no advice I've gotten has been close to what I need.
CodePudding user response:
One way you can do it using lapply and sapply
#Get Matches for column name and country variable
matches <- lapply(colnames(df[-(1:2)]), \(x) df$country %in% x)
#Get the id for each match
ids <- lapply(matches, \(x) df$id[x])
#add 1 to each
df[-(1:2)] <- sapply(ids, \(x) (df$id %in% x))
or in one go
df[-(1:2)] <- sapply(lapply(lapply(colnames(df[-(1:2)]),
\(x) df$country %in% x),
\(x) df$id[x]),
\(x) (df$id %in% x))
id country US JP CH KR BE SP
1 1 US 1 1 0 0 0 0
2 1 JP 1 1 0 0 0 0
3 3 CH 1 0 1 0 0 0
4 3 US 1 0 1 0 0 0
5 5 KR 0 0 0 1 0 0
6 6 KR 0 0 0 1 0 0
CodePudding user response:
Issue in the OP's code seems to be related to the class of 'country' which is character. We may need to convert to factor before doing the model.matrix and include the levels based on the column names in 'data2'. Also, wrapping factor on the 'data2$country' will have only the values from that column (as some of them are still missing based on the column names)
data2$country <- factor(data2$country, levels = names(data2)[-(1:2)])
m1 <- model.matrix(~country - 1, data2)
m1[] <- ave(c(m1), data2$id[row(m1)], col(m1), FUN = max)
data2[levels(data2$country)] <- m1
-output
> data2
id country US JP CH KR BE SP
1 1 US 1 1 0 0 0 0
2 1 JP 1 1 0 0 0 0
3 3 CH 1 0 1 0 0 0
4 3 US 1 0 1 0 0 0
5 5 KR 0 0 0 1 0 0
6 6 KR 0 0 0 1 0 0
CodePudding user response:
Here we first group by
id, then assign 1 to to each column that has same name as the row in country.Next step is a trick: we use
fillto get 1 for each row of 1 in the groupNext we bring back the NA to 0
library(dplyr)
df %>%
group_by(id) %>%
mutate(across(-country, ~case_when(country == cur_column() ~ 1))) %>%
fill(-country, .direction = "updown") %>%
mutate(across(-country, ~ifelse(is.na(.), 0, .))) %>%
ungroup()
output:
id country US JP CH KR BE SP
<dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 US 1 1 0 0 0 0
2 1 JP 1 1 0 0 0 0
3 3 CH 1 0 1 0 0 0
4 3 US 1 0 1 0 0 0
5 5 KR 0 0 0 1 0 0
6 6 KR 0 0 0 1 0 0
CodePudding user response:
Here is another tidyverse option, where we put into long form, then group by the id, then if the column name is in the country column, then change to a 1. Then, we can pivot back to the wide form.
library(tidyverse)
df %>%
pivot_longer(-c(id, country)) %>%
group_by(id) %>%
mutate(value = ifelse(name %in% country, 1, 0)) %>%
ungroup %>%
pivot_wider(names_from = "name", values_from = "value")
Output
id country US JP CH KR BE SP
<dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 US 1 1 0 0 0 0
2 1 JP 1 1 0 0 0 0
3 3 CH 1 0 1 0 0 0
4 3 US 1 0 1 0 0 0
5 5 KR 0 0 0 1 0 0
6 6 KR 0 0 0 1 0 0