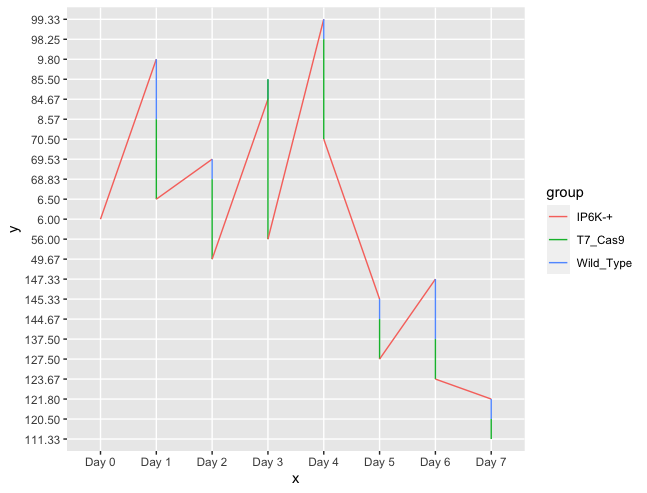
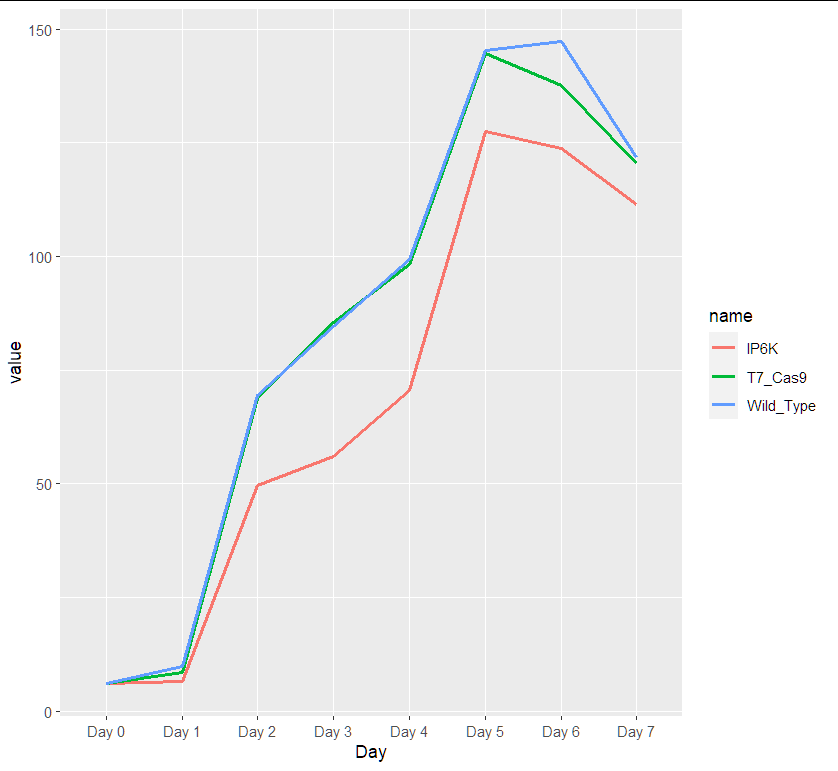
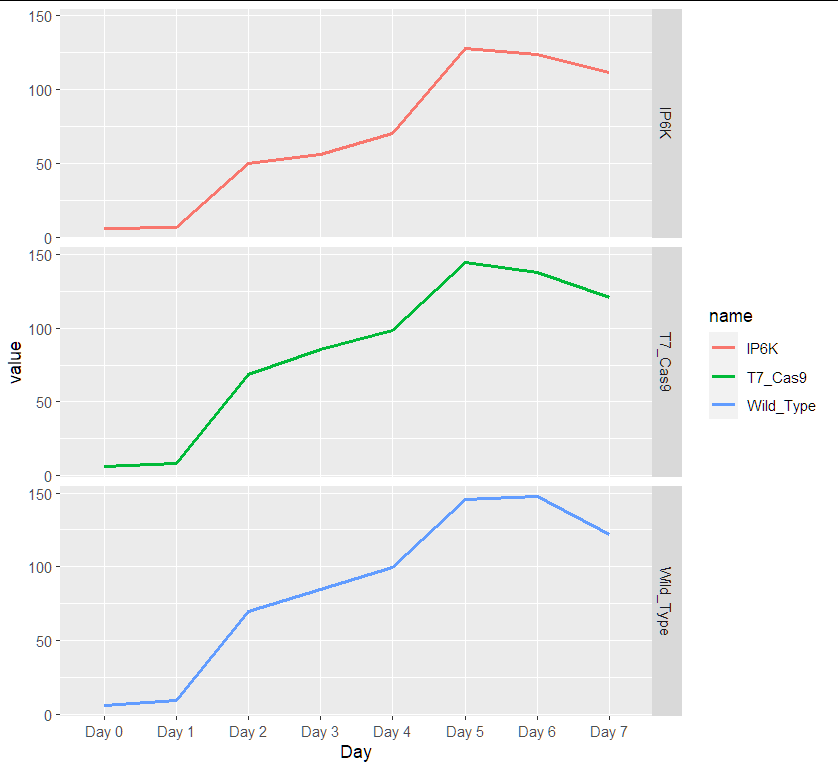
Please help me. I have the following data in R: I have values of three groups of organisms from day 0 to day 7 which represent the mean of populations for these groups for each day.
Here is my data:
However, what I would like to have is:
CodePudding user response:
Are you looking for such a solution:
library(tidyverse)
df %>%
pivot_longer(-Day) %>%
ggplot(aes(x = Day, y = value, group=name, color = name))
geom_line(size=1)
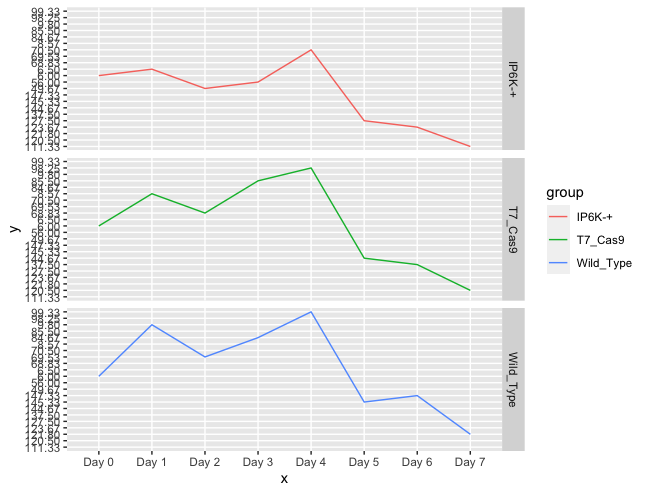
AND with facet
library(tidyverse)
df %>%
pivot_longer(-Day) %>%
ggplot(aes(x = Day, y = value, group=name, color = name))
geom_line(size=1)
facet_grid(name ~ .)
data:
df <- structure(list(Day = c("Day 0", "Day 1", "Day 2", "Day 3", "Day 4",
"Day 5", "Day 6", "Day 7"), Wild_Type = c(6, 9.8, 69.53, 84.67,
99.33, 145.33, 147.33, 121.8), T7_Cas9 = c(6, 8.57, 68.83, 85.5,
98.25, 144.67, 137.5, 120.5), IP6K = c(6, 6.5, 49.67, 56, 70.5,
127.5, 123.67, 111.33)), class = "data.frame", row.names = c(NA,
-8L))
CodePudding user response:
Try:
scale_y_continuous(breaks = seq(1, 7, 1), limits = c(0, 7), labels = c())
I guess you could play around with the labels argument; also not sure about your data, but some transformations (eg log) may help to separate the data better!