I want to calculate the mean and standard deviation for subgroups every column in my dataset. The membership of the subgroups is based on the values in the column of interest and these subgroups are specific to each column of interest.
# Example data
set.seed(1)
library(data.table)
df <- data.frame(baseline = runif(100), `Week0_12` = runif(100), `Week12_24` = runif(100))
So for column Baseline, a row may be assigned to another subgroup than for column Week0_12.
I can of course create these 'subgroup columns' manually for each column and then calculate the statistics for each column by column subgroup:
df$baseline_subgroup <- ifelse(df$baseline < 0.2, "subgroup_1", "subgroup_2")
df <- as.data.table(df)
df[, .(mean = mean(baseline), sd = sd(baseline)), by = baseline_subgroup]
Giving this output:
baseline_subgroup mean sd
1: subgroup_2 0.58059314 0.22670071
2: subgroup_1 0.09793105 0.05317809
Doing this for every column separately is too much repetition, especially given that I have many columns my actual data.
df$Week0_12_subgroup <- ifelse(df$Week0-12 < 0.2, "subgroup_1", "subgroup_2")
df[, .(mean = mean(Week0_12), sd = sd(Week0_12 )), by = Week0_12_subgroup ]
df$Week12_24_subgroup <- ifelse(df$Week0-12 < 0.2, "subgroup_1", "subgroup_2")
df[, .(mean = mean(Week12_24), sd = sd(Week12_24)), by = Week12_24_subgroup ]
What is a more elegant approach to do this?
CodePudding user response:
Here's a tidyverse method that gives an easy-to-read and easy-to-plot output:
library(tidyverse)
set.seed(1)
df <- data.frame(baseline = runif(100),
`Week0_12` = runif(100),
`Week12_24` = runif(100))
df2 <- df %>%
summarize(across(everything(), list(mean_subgroup1 = ~mean(.x[.x < 0.2]),
sd_subgroup1 = ~sd(.x[.x < 0.2]),
mean_subgroup2 = ~mean(.x[.x > 0.2]),
sd_subgroup2 = ~sd(.x[.x > 0.2])))) %>%
pivot_longer(everything(), names_pattern = '^(.*)_(.*)_(.*$)',
names_to = c('time', 'measure', 'subgroup')) %>%
pivot_wider(names_from = measure, values_from = value)
df2
#> # A tibble: 6 x 4
#> time subgroup mean sd
#> <chr> <chr> <dbl> <dbl>
#> 1 baseline subgroup1 0.0979 0.0532
#> 2 baseline subgroup2 0.581 0.227
#> 3 Week0_12 subgroup1 0.117 0.0558
#> 4 Week0_12 subgroup2 0.594 0.225
#> 5 Week12_24 subgroup1 0.121 0.0472
#> 6 Week12_24 subgroup2 0.545 0.239
ggplot(df2, aes(time, mean, group = subgroup))
geom_errorbar(aes(ymin = mean - sd, ymax = mean sd, color = subgroup),
width = 0.1)
geom_point()
theme_minimal(base_size = 16)
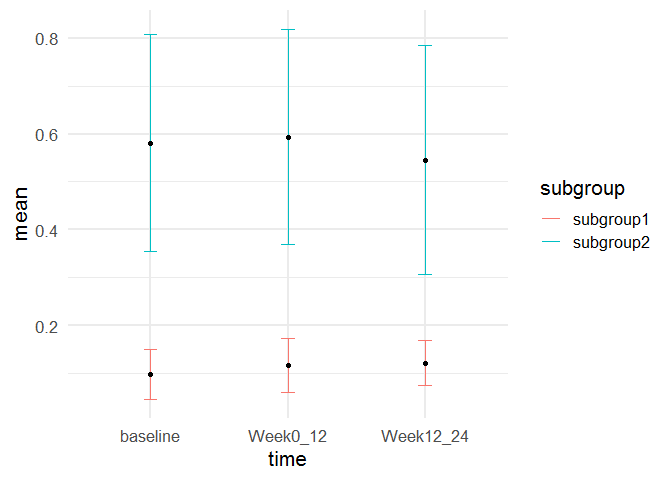
Created on 2022-07-14 by the reprex package (v2.0.1)
CodePudding user response:
You could use apply to apply a subgroup function across each column
i. e.
# list to house dfs of summary statistics
summaries <- list()
subgroup <- function(x){
# x is the column that we are interested in
df$current_subgroup<- ifelse(x < 0.2, "subgroup_1", "subgroup_2")
library(data.table)
df <- as.data.table(df)
summaries.append(df[, .(mean = mean(baseline), sd = sd(baseline)), by = baseline_subgroup])
}
# MARGIN = 2 applies across columns
apply(df, 2, subgroup)
CodePudding user response:
You can create a custom function and apply it using .SD, i.e.
library(data.table)
f1 <- function(x){
i_mean <- mean(x);
i_sd <- sd(x);
list(Avg = i_mean, standard_dev = i_sd)
}
setDT(df)[, unlist(lapply(.SD, f1), recursive = FALSE), by = baseline_subgroup][]
baseline_subgroup baseline.Avg baseline.standard_dev Week0.12.Avg Week0.12.standard_dev Week12.24.Avg Week12.24.standard_dev
1: subgroup_2 0.5950020 0.22556590 0.5332555 0.2651810 0.5467046 0.2912027
2: subgroup_1 0.1006693 0.04957005 0.5947161 0.2645519 0.5137543 0.3213723
