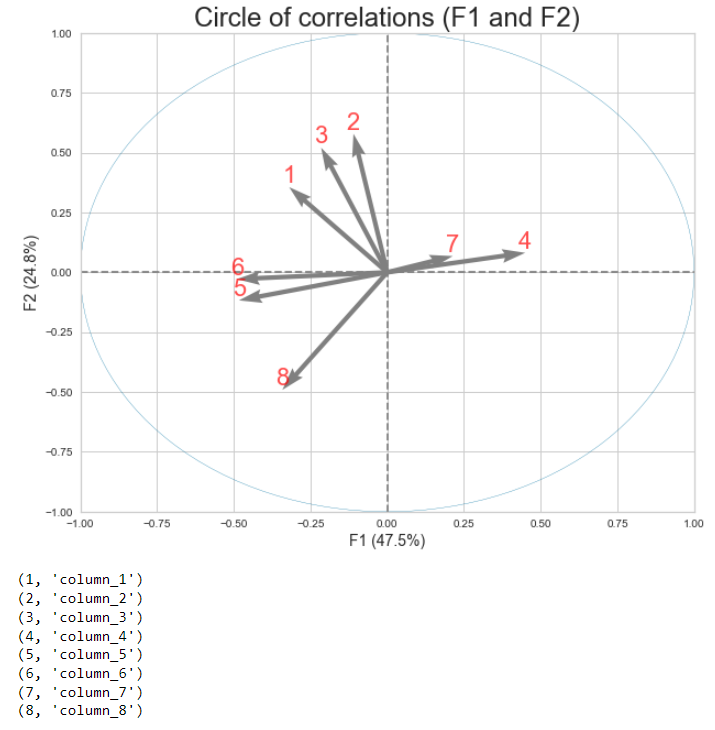
I have a function that allows me to display the circle of correlations of my pca.
The problem with this function is that the labels of my variables (column names) prevent me from reading my results correctly. To overcome this problem, I have to insert a line of code before my function to associate numbers with the labels of the variables (of the df used to make my pca):
n_labels = [value for value in range(1, (len(df.columns) 1))]
I tried unsuccessfully to insert this line in my function:
import matplotlib.pyplot as plt
from matplotlib.collections import LineCollection
import numpy as np
import pandas as pd
def display_circles(pcs,
n_comp,
pca,
axis_ranks,
labels=None,
label_rotation=0,
lims=None):
for d1, d2 in axis_ranks: # We display the first 3 factorial planes, so the first 6 components
if d2 < n_comp:
# figure initialization
fig, ax = plt.subplots(figsize=(10, 8))
# determination of graph limits
if lims is not None:
xmin, xmax, ymin, ymax = lims
elif pcs.shape[1] < 30:
xmin, xmax, ymin, ymax = -1, 1, -1, 1
else:
xmin, xmax, ymin, ymax = min(pcs[d1, :]), max(pcs[d1, :]), min(
pcs[d2, :]), max(pcs[d2, :])
# arrow display
# if there are more than 30 arrows, the triangle is not displayed at their end
if pcs.shape[1] < 30:
plt.quiver(np.zeros(pcs.shape[1]),
np.zeros(pcs.shape[1]),
pcs[d1, :],
pcs[d2, :],
angles='xy',
scale_units='xy',
scale=1,
color="grey")
else:
lines = [[[0, 0], [x, y]] for x, y in pcs[[d1, d2]].T]
ax.add_collection(
LineCollection(lines, axes=ax, alpha=.1, color='black'))
# display of variable names
if labels is not None:
for i, (x, y) in enumerate(pcs[[d1, d2]].T):
if x >= xmin and x <= xmax and y >= ymin and y <= ymax:
plt.text(x,
y,
labels[i],
fontsize='22',
ha='center',
va='bottom',
rotation=label_rotation,
color="red",
alpha=0.7)
# circle display
circle = plt.Circle((0, 0), 1, facecolor='none', edgecolor='b')
plt.gca().add_artist(circle)
# setting graph limits
plt.xlim(xmin, xmax)
plt.ylim(ymin, ymax)
# display of horizontal and vertical lines
plt.plot([-1, 1], [0, 0], color='grey', ls='--')
plt.plot([0, 0], [-1, 1], color='grey', ls='--')
# names of the axes, with the percentage of inertia explained
plt.xlabel('F{} ({}%)'.format(
d1 1, round(100 * pca.explained_variance_ratio_[d1], 1)),
fontsize=14)
plt.ylabel('F{} ({}%)'.format(
d2 1, round(100 * pca.explained_variance_ratio_[d2], 1)),
fontsize=14)
plt.title("Circle of correlations (F{} and F{})".format(
d1 1, d2 1),
size=24)
plt.show()
This is how I call my function:
import pandas as pd
from sklearn import decomposition, preprocessing
# Here a dataset for the example:
column_1 = [1, 2, 3, 4, 5, 6, 7 ,8]
column_2 = [4, 2, 9, 23, 3, 52, 41, 4]
column_3 = [9, 8, 7, 6, 6, 9, 24, 11]
column_4 = [45, 36, 74, 35, 29, 45, 29, 39]
column_5 = [35, 84, 3, 54, 68, 78, 65, 97]
column_6 = [24, 96, 7, 54, 67, 69, 88, 95]
column_7 = [5, 39, 72, 42, 22, 41, 24, 41]
column_8 = [30, 98, 8, 67, 68, 41, 27, 87]
df = pd.DataFrame({'column_1': column_1,
'column_2': column_2,
'column_3': column_3,
'column_4': column_4,
'column_5': column_5,
'column_6': column_6,
'column_7': column_7,
'column_8': column_8})
pca_data = preprocessing.scale(df)
pca = decomposition.PCA(n_components = 8)
pca.fit(pca_data)
# We set the number of components
n_comp = 2
# Recovery of the components of the pca object
pcs = pca.components_
# We label a number to each column name
n_labels = [value for value in range(1, (len(df.columns) 1))]
display_circles(pcs, n_comp, pca, [(0, 1), (0, 2)], labels=n_labels)
for element in zip(n_labels, df.columns):
print(element)
Edit 1: that i would like (UPD: with the answer of @Stef -Thanks you very much and congratulations for this solution-)
it's almost perfect but the problem is when I use this function:
n_comp = 3
pcs = pca.components_
# I always have to write this line to get a label number
n_labels=[value for value in range(1,(len(list_candidates.columns) 1))]
display_circles(pcs, n_comp, pca, [(0, 1), (0, 2)], labels=n_labels)
on my real dataframe, this throws me two problems:
- I still have to include the line
n_labels=[value for value in range(1,(len(list_candidates.columns) 1))]
to obtain a label number instead of the name of my variables.
- I get the error message "NameError: name 'df' is not defined" when running
display_circles(pcs, n_comp, pca, [(0, 1), (0, 2)], labels=n_labels)
So I'm looking to define my display_circles() function so that when I set the labels="name_of_the_df" argument it returns me the same result as
n_labels=[value for value in range(1,(len("name_of_the_df".columns) 1))]
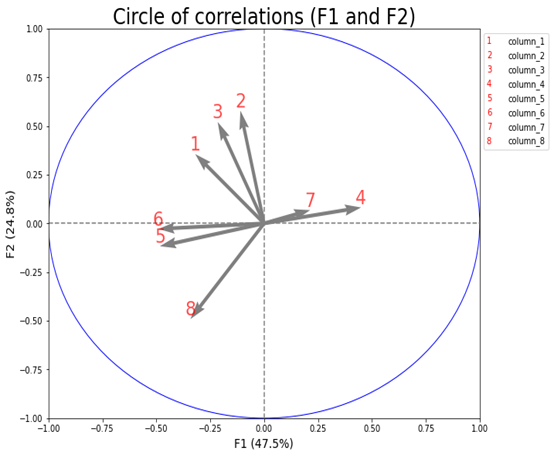
plus a plt.legend() like the one made by @Steph (thanks)
To get this (desired) result:
I also have to modify "name_of_the_df" in the function definition:
#legend
plt.legend(n_labels,
candidate_list.columns,
handler_map={int: IntHandler()},
bbox_to_anchor=(1, 1))
CodePudding user response:
Full example as per comment below and edited question:
import matplotlib.pyplot as plt
from matplotlib.collections import LineCollection
from matplotlib.text import Text
import numpy as np
import pandas as pd
from sklearn import decomposition, preprocessing
class IntHandler:
def legend_artist(self, legend, orig_handle, fontsize, handlebox):
x0, y0 = handlebox.xdescent, handlebox.ydescent
text = Text(x0, y0, str(orig_handle), color='red')
handlebox.add_artist(text)
return text
def display_circles(pcs,
n_comp,
pca,
axis_ranks,
labels=None,
label_rotation=0,
lims=None):
if labels == None:
labels = range(1, len(pca.feature_names_in_) 1)
for d1, d2 in axis_ranks: # We display the first 3 factorial planes, so the first 6 components
if d2 < n_comp:
# figure initialization
fig, ax = plt.subplots(figsize=(10, 8))
# determination of graph limits
if lims is not None:
xmin, xmax, ymin, ymax = lims
elif pcs.shape[1] < 30:
xmin, xmax, ymin, ymax = -1, 1, -1, 1
else:
xmin, xmax, ymin, ymax = min(pcs[d1, :]), max(pcs[d1, :]), min(
pcs[d2, :]), max(pcs[d2, :])
# arrow display
# if there are more than 30 arrows, the triangle is not displayed at their end
if pcs.shape[1] < 30:
plt.quiver(np.zeros(pcs.shape[1]),
np.zeros(pcs.shape[1]),
pcs[d1, :],
pcs[d2, :],
angles='xy',
scale_units='xy',
scale=1,
color="grey")
else:
lines = [[[0, 0], [x, y]] for x, y in pcs[[d1, d2]].T]
ax.add_collection(
LineCollection(lines, axes=ax, alpha=.1, color='black'))
# display of variable names
if labels is not None:
for i, (x, y) in enumerate(pcs[[d1, d2]].T):
if x >= xmin and x <= xmax and y >= ymin and y <= ymax:
plt.text(x,
y,
labels[i],
fontsize='22',
ha='center',
va='bottom',
rotation=label_rotation,
color="red",
alpha=0.7,
)
# circle display
circle = plt.Circle((0, 0), 1, facecolor='none', edgecolor='b')
plt.gca().add_artist(circle)
# setting graph limits
plt.xlim(xmin, xmax)
plt.ylim(ymin, ymax)
# display of horizontal and vertical lines
plt.plot([-1, 1], [0, 0], color='grey', ls='--')
plt.plot([0, 0], [-1, 1], color='grey', ls='--')
# names of the axes, with the percentage of inertia explained
plt.xlabel('F{} ({}%)'.format(
d1 1, round(100 * pca.explained_variance_ratio_[d1], 1)),
fontsize=14)
plt.ylabel('F{} ({}%)'.format(
d2 1, round(100 * pca.explained_variance_ratio_[d2], 1)),
fontsize=14)
plt.title("Circle of correlations (F{} and F{})".format(
d1 1, d2 1),
size=24)
plt.legend(labels,
pca.feature_names_in_,
handler_map={int: IntHandler()},
bbox_to_anchor=(1,1))
plt.show()
# Here a dataset for the example:
column_1 = [1, 2, 3, 4, 5, 6, 7 ,8]
column_2 = [4, 2, 9, 23, 3, 52, 41, 4]
column_3 = [9, 8, 7, 6, 6, 9, 24, 11]
column_4 = [45, 36, 74, 35, 29, 45, 29, 39]
column_5 = [35, 84, 3, 54, 68, 78, 65, 97]
column_6 = [24, 96, 7, 54, 67, 69, 88, 95]
column_7 = [5, 39, 72, 42, 22, 41, 24, 41]
column_8 = [30, 98, 8, 67, 68, 41, 27, 87]
df = pd.DataFrame({'column_1': column_1,
'column_2': column_2,
'column_3': column_3,
'column_4': column_4,
'column_5': column_5,
'column_6': column_6,
'column_7': column_7,
'column_8': column_8})
pca_data = preprocessing.scale(df)
pca = decomposition.PCA(n_components = 8)
pca.fit(pd.DataFrame(pca_data, columns=df.columns))
# We set the number of components
n_comp = 2
# Recovery of the components of the pca object
pcs = pca.components_
display_circles(pcs, n_comp, pca, [(0, 1), (0, 2)])