I have a netCDF file downloaded from the 
It contains annual temperature anomaly value relative to certain time period.
I want to plot the annual temperature values for four years in the form of subplots. I use the following code:
fig, axs = plt.subplots(2, 2, figsize = (10, 10))
fig.suptitle("Temperature anomaly in the end of last four decades\n relative to 1951-1980")
def get_plot(year, i, j, k):
ax = fig.add_subplot(2, 2, k,
projection = ccrs.PlateCarree())
ax.add_feature(NaturalEarthFeature('cultural', 'admin_0_countries', '10m'),
facecolor='none', edgecolor='black')
ax.set_extent([-150, 150, -55, 85])
xr_df_sub = anomaly.loc[f"{year}-12-31"]
ax.set_title(f"{year}")
ax.set_axis_off()
xr.plot.imshow(xr_df_sub,
ax = ax,
add_labels = False,
vmin = -4, vmax = 4,
cmap = "coolwarm",
add_colorbar = False,
interpolation = "bicubic")
axs[0, 0] = get_plot(1990, 0, 0, 1)
axs[0, 1] = get_plot(2000, 0, 1, 2)
axs[1, 0] = get_plot(2010, 0, 1, 3)
axs[1, 1] = get_plot(2020, 0, 1, 4)
# add colorbar
cax = fig.add_axes([0.92, 0.15, 0.01, 0.7]) #[left, bottom, width, height]
sm = plt.cm.ScalarMappable(cmap='coolwarm',
norm=plt.Normalize(vmin= -4, vmax= 4))
# fake up the array of the scalar mappable.
sm._A = []
lgd=fig.colorbar(sm, cax=cax, extend = "both"
).set_label("°C", rotation=0,y=1.1, labelpad= -35)
plt.show()
I created a function called get_plot(year, i, j, k) for each subplot. i, j refer to row and column number and k refer to the index number of the subplot. The function to plot the temperature anomaly for given year.
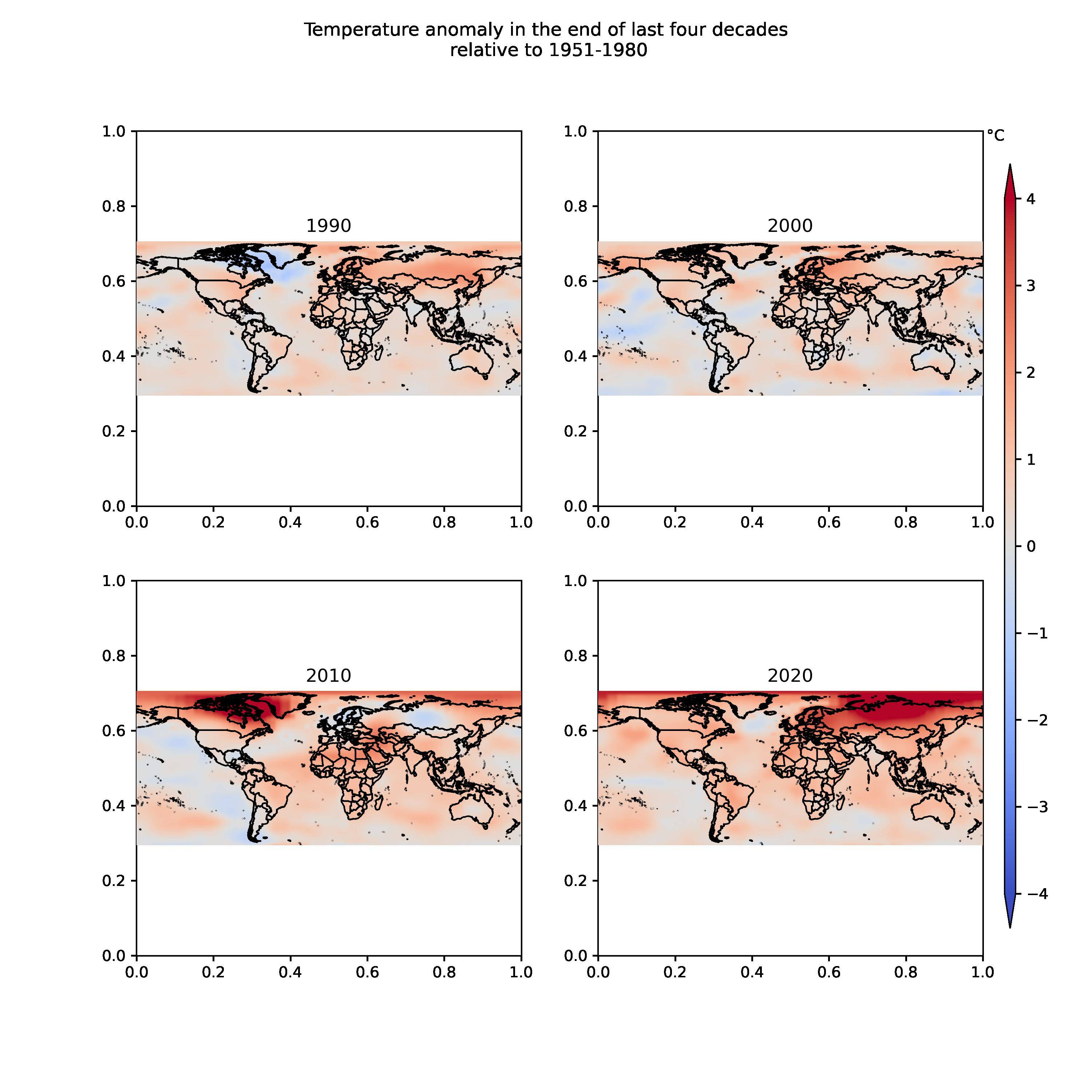
This is the desired plot, however, I am getting outer black frames for each subplot, which I want to remove. I used following code:
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.spines['bottom'].set_visible(False)
ax.spines['left'].set_visible(False)
However, it does not change anything. I also tried
fig.patch.set_visible(False)
It also does not change anything.
How can I remove the outer borders with 0 to 1 in x-axis and y-axis for each subplot? Also, I'd like to get advice on bringing subplots closer to each other?
CodePudding user response:
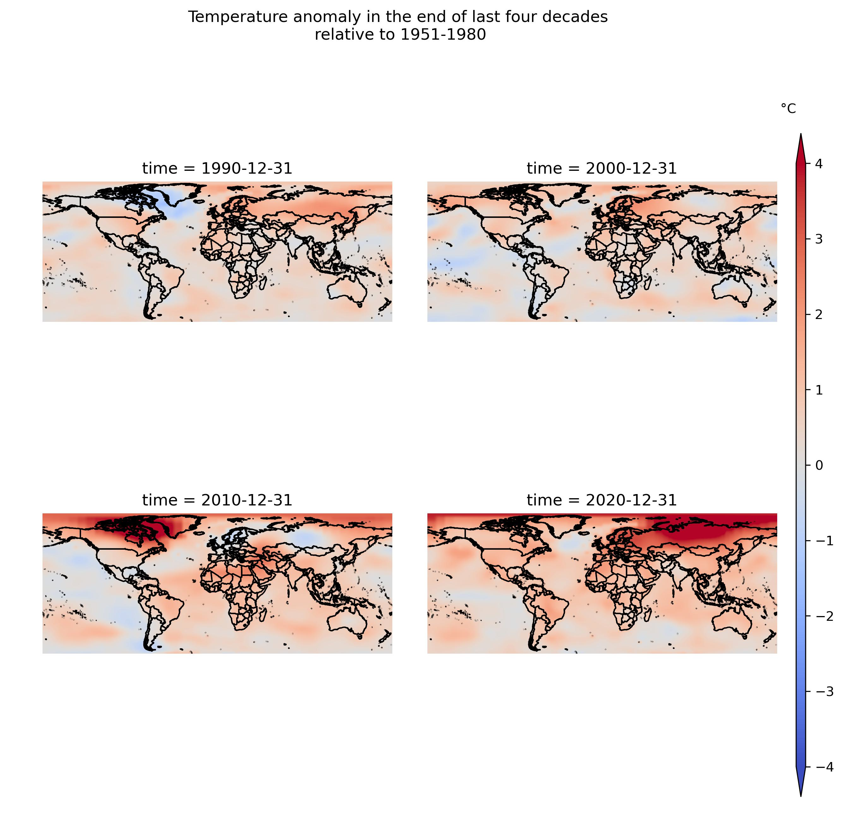
Based on suggestion of Dahn, I figured out that I had used plt.subplot() and again used fig.add_subplot() twice.
I solved the problem as follows:
fig = plt.figure(figsize = (10, 10))
fig.suptitle("Temperature anomaly in the end of last four decades\n relative to 1951-1980", y = 0.8)
plt.subplots_adjust(bottom = 0.3, top = 0.7, wspace=0.1, hspace= 0.25)
def get_plot(year, i, j, k):
ax = fig.add_subplot(2, 2, k,
projection = ccrs.PlateCarree())
ax.add_feature(NaturalEarthFeature('cultural', 'admin_0_countries', '10m'),
facecolor='none', edgecolor='black')
ax.set_extent([-150, 150, -55, 85])
xr_df_sub = anomaly.loc[f"{year}-12-31"]
ax.set_title(f"{year}")
ax.axis("off")
ax = xr.plot.imshow(xr_df_sub,
add_labels = True,
vmin = -4, vmax = 4,
cmap = "coolwarm",
add_colorbar = False,
interpolation = "bicubic",
)
return ax
ax[0, 0] = get_plot(1990, 0, 0, 1)
ax[0, 1] = get_plot(2000, 0, 1, 2)
ax[1, 0] = get_plot(2010, 0, 1, 3)
ax[1, 1] = get_plot(2020, 0, 1, 4)
# add colorbar
cax = fig.add_axes([0.92, 0.3, 0.02, 0.5]) #[left, bottom, width, height]
sm = plt.cm.ScalarMappable(cmap='coolwarm',
norm=plt.Normalize(vmin= -4, vmax= 4))
# fake up the array of the scalar mappable.
sm._A = []
lgd=fig.colorbar(sm, cax=cax, extend = "both"
).set_label("°C", rotation=0,y=1.1, labelpad= -35)
plt.show()