I tried to implement the vmax option with np.clip however when I compare the built-in version and my implementation I get different results.
import numpy as np
import matplotlib.pyplot as plt
from PIL import Image
np.random.seed(0)
size = 900
percentile_clip = 30
data_source = np.random.rand(size, size)
data_source = data_source * (255/data_source.max())
data_source = data_source.astype('uint8')
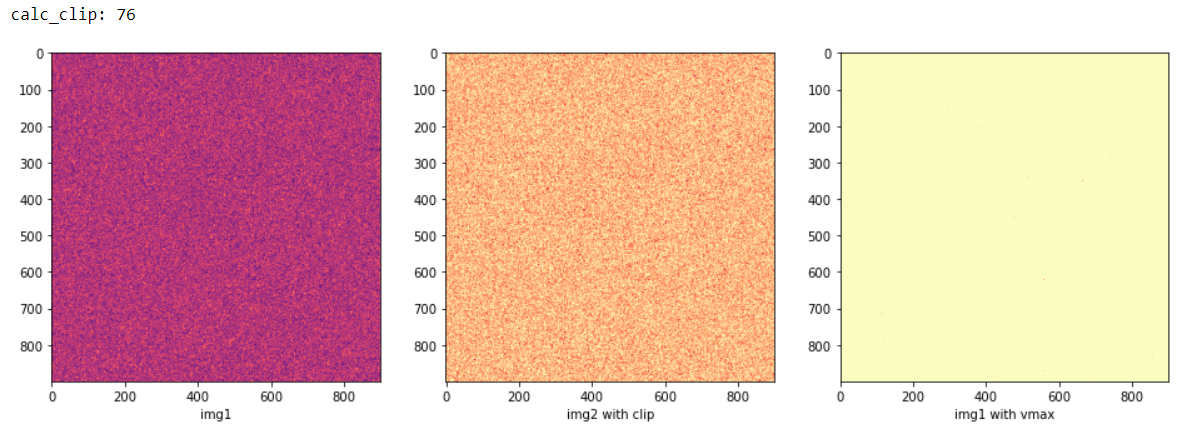
calc_clip = int(np.percentile(data_source, percentile_clip))
img1 = Image.fromarray(data_source, mode='L')
img2 = Image.fromarray(np.clip(data_source,0,calc_clip), mode='L')
fig, axs = plt.subplots(1, 3,figsize=(16,8))
axs[0].imshow(img1,cmap='magma')
axs[0].set_xlabel('img1')
axs[1].imshow(img2,cmap='magma')
axs[1].set_xlabel('img2 with clip')
axs[2].imshow(img1,cmap='magma',vmin = 0, vmax = calc_clip)
axs[2].set_xlabel('img1 with vmax')
print('calc_clip:',calc_clip)
 On the middle and right plot I should get similar results. If I modify the
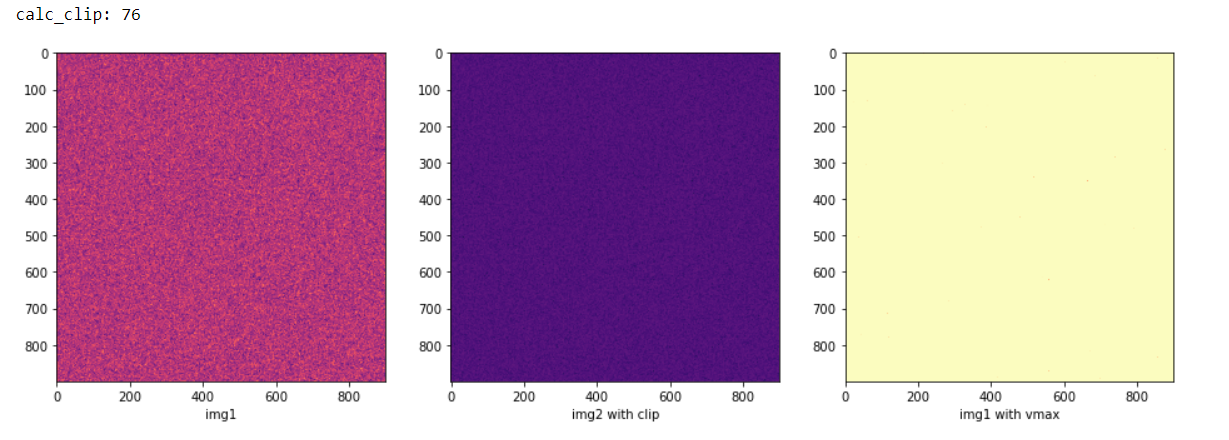
On the middle and right plot I should get similar results. If I modify the size to a smaller number e.g. 10 then it works, but with larger values the results are completely different. Is this in connection with storing data as uint8?
I also tried manually adding vmin = 0, vmax = 255 to axs[0] and axs[\1] as well, then the results became even stranger.
CodePudding user response:
This is because when size=900, all your images are aliased because you have nowhere near as many pixels on the screen as you have in the image, so Matplotlib has to do something....
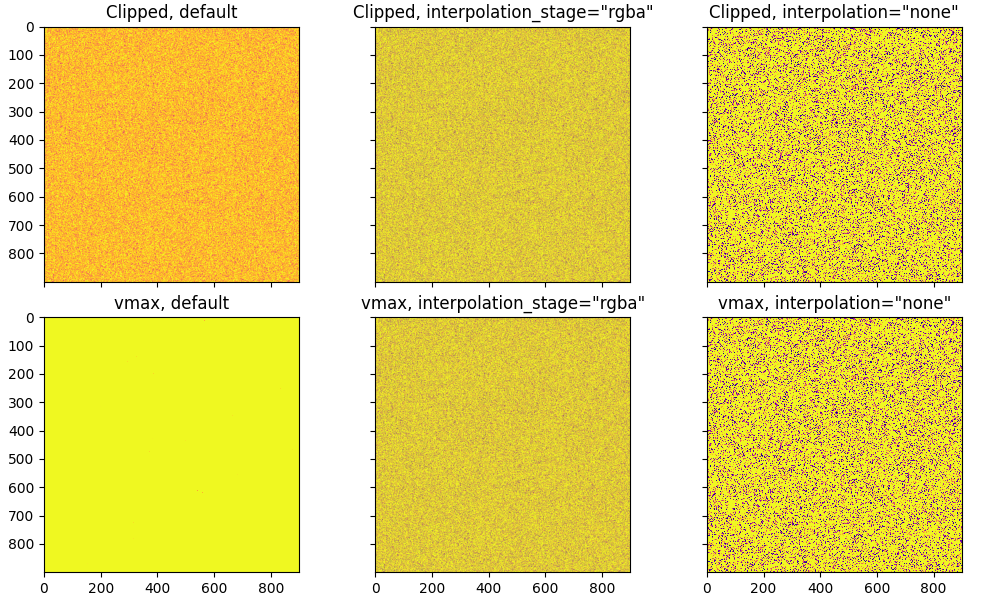
By default matplotlib will smooth the data, and then apply the norm. The problem with that is that the high values that you are trying to clip leak into the smoothing and lead to the vmax version (third subplot) having "brighter" colors than the clipped version.
If you want a reasonable view of your data, you can increase the dpi and view the resulting png in an image viewer.
You can also, with Matplotlib set the interpolation_stage to rgba, and then the anti-aliasing smoothing will happen in RGBA space rather than data space. Or you can have an aliased image with interpolation='none':
from configparser import Interpolation
import numpy as np
import matplotlib.pyplot as plt
from PIL import Image
np.random.seed(0)
size = 900
percentile_clip = 30
data_source = np.random.rand(size, size)
data_source = data_source * (255/data_source.max())
data_source = data_source.astype('uint8')
calc_clip = int(np.percentile(data_source, percentile_clip))
img1 = Image.fromarray(data_source, mode='L')
img2 = Image.fromarray(np.clip(data_source,0,calc_clip), mode='L')
fig, axs = plt.subplots(2, 3, figsize=(10, 6), sharex=True, sharey=True, constrained_layout=True)
axs[0, 0].imshow(img2, cmap='plasma')
axs[0, 0].set_title('Clipped, default')
axs[0, 1].imshow(img2, cmap='plasma', interpolation_stage='rgba')
axs[0, 1].set_title('Clipped, interpolation_stage="rgba"')
axs[0, 2].imshow(img2, cmap='plasma', interpolation='none')
axs[0, 2].set_title('Clipped, interpolation="none"')
axs[1, 0].imshow(img1, cmap='plasma', vmin=0, vmax=calc_clip)
axs[1, 0].set_title('vmax, default')
axs[1, 1].imshow(img1, cmap='plasma', interpolation_stage='rgba', vmin=0, vmax=calc_clip)
axs[1, 1].set_title('vmax, interpolation_stage="rgba"')
axs[1, 2].imshow(img1, cmap='plasma', interpolation='none', vmin=0, vmax=calc_clip)
axs[1, 2].set_title('vmax, interpolation="none"')
plt.show()
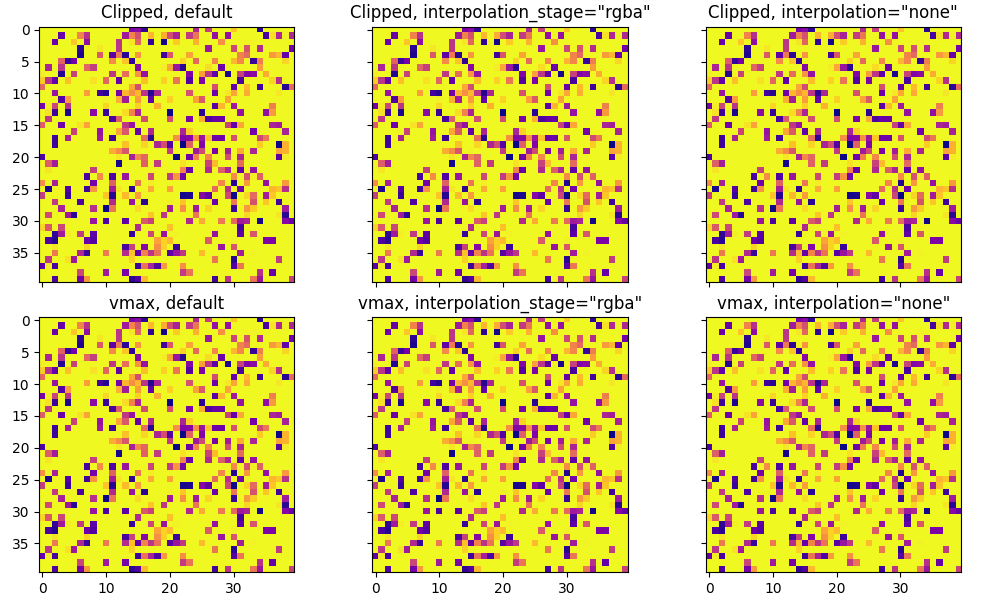
If you aren't aliasing your image (the size is smaller) these are all identical: eg with size=40:
Note that interpolation='none' can give pretty misleading results if you care about those individual pixels - sometimes spikes will show up, sometimes they will be completely obscured. A lot of advice seems to be to use interpolation='none', but do so only at your own risk.
For more discussion, please see https://matplotlib.org/stable/gallery/images_contours_and_fields/image_antialiasing.html