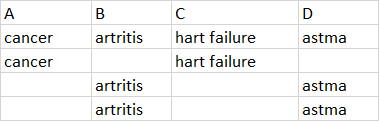
I have several columns that contain specific diseases. Here an example of a piece of it:
I want to make all possible combinations so I can check which combination of diseases mostly occur. So I want to make all combinations of 2 columns (A&B, A&C, A&D, B&C, B&D, C&D), but also combinations of 3 and 4 columns (A&B&C, B&C&D and so on). I have the following script for this:
from itertools import combinations
df.join(pd.concat({'_'.join(x): df[x[0]].str.cat(df[list(x[1:])].astype(str),
sep='')
for i in (2, 3, 4)
for x in combinations(df, i)}, axis=1))
But that generates a lot of extra columns in my dataset, and I still haven't got the frequencies of all combinations. This is the output that I would like to get:
What script can I use for this?
CodePudding user response:
Use DataFrame.stack with aggregate join and last count by Series.value_counts:
s = df.stack().groupby(level=0).agg(','.join).value_counts()
print (s)
artritis,asthma 2
cancer,artritis,heart_failure,asthma 1
cancer,heart_failure 1
dtype: int64
If need 2 columns DataFrame:
df = s.rename_axis('vals').reset_index(name='count')
print (df)
vals count
0 artritis,asthma 2
1 cancer,artritis,heart_failure,asthma 1
2 cancer,heart_failure 1
CodePudding user response:
You can create a pivot table
def index_agg_fn(x):
x = [e for e in x if e != '']
return ','.join(x)
df = pd.DataFrame({'A': ['cancer', 'cancer', None, None],
'B': ['artritis', None, 'artritis', 'artritis'],
'C': ['heart_failure', 'heart_failure', None, None],
'D': ['asthma', None, 'asthma', 'asthma']})
df['count'] = 1
ptable = pd.pivot_table(df.fillna(''), index=['A', 'B', 'C', 'D'], values=['count'], aggfunc='sum')
ptable.index = list(map(index_agg_fn, ptable.index))
print(ptable)
Result
count
artritis,asthma 2
cancer,heart_failure 1
cancer,artritis,heart_failure,asthma 1