#splitting dataset into train, val, and test into 60-20-20
features = df.drop('quality', axis = 1)
labels = df['quality']
X_train, X_test, y_train, y_test = train_test_split(features, labels, test_size=0.4, random_state=42)
X_test, X_val, y_test, y_val = train_test_split(X_test, y_test, test_size=0.5, random_state=42)
X_train.to_csv('train_features.csv', index=False)
X_val.to_csv('val_features.csv', index=False)
X_test.to_csv('test_features.csv', index=False)
y_train.to_csv('train_labels.csv', index=False)
y_val.to_csv('val_labels.csv', index=False)
y_test.to_csv('test_labels.csv', index=False)
from sklearn.ensemble import RandomForestRegressor
rfModel = RandomForestRegressor(n_estimators=500)
rfModel.fit(X_train, np.log1p(y_train))
preds2 = rfModel.predict(X_test)
The y_train.info() is:
count 815
unique 2
top 0
freq 704
Name: quality, dtype: int64
The y_train.head() is:
635 0
908 0
1578 0
245 0
1451 1
Name: quality, dtype: category
Categories (2, int64): [0 < 1]
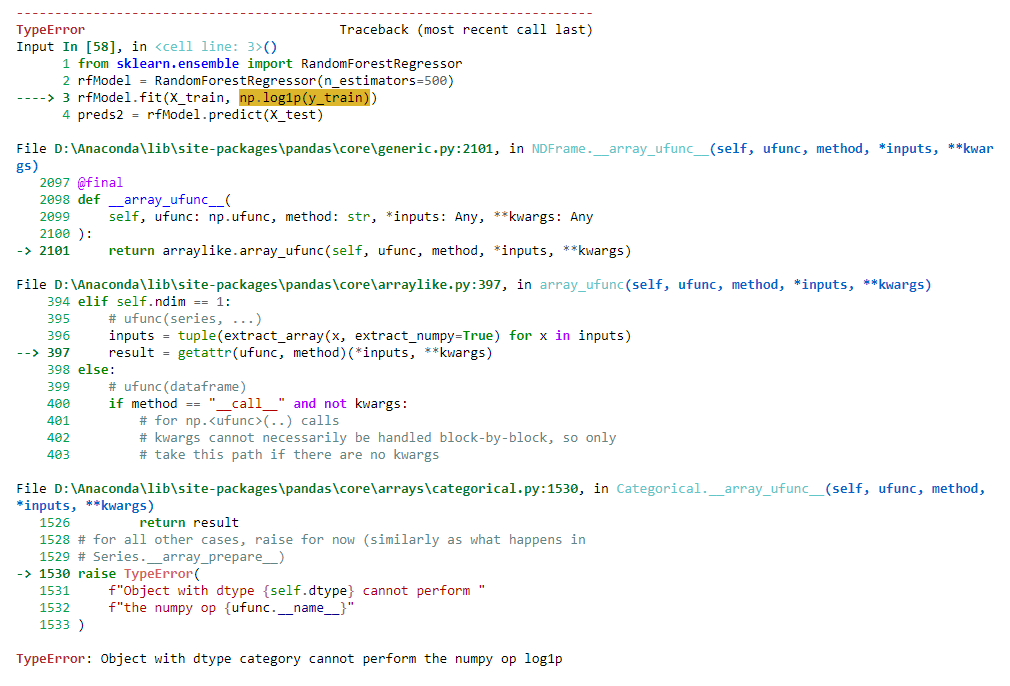
I am clueless about why it is error, I could use np.log1p(1e-99) with other but not this one.
The error info is as below:
CodePudding user response:
The issue is that you attempt to apply the logarithm to an array-like object that is of type categorical rather than int (or np.int64 etc.).
In particular, the error can be produced by certain Pandas columns when a class label is encoded as a category rather than a mere integer. This is usually the case, we the class label of the endogenous variable is encoded as for y_train and revealed by your command y_train.head(). Here, is a minimum viable example how it pops up.
s = pd.Series([1,2,3,4], dtype="category")
np.log1p(s) # TypeError: Object with dtype category cannot perform the numpy op log1p
However, converting the type to float (or int etc.) solves the problem, e.g.
s = pd.Series([1,2,3,4], dtype="category")
s = s.astype(float)
np.log1p(s) # no error/warning
