I would like to know how to create a new column with conditionals values.
I have a dataframe called 'df' with 4 columns : ID of the person , Question, title of the question, and Answer . I would like to add a column 'TEST' depending on the answer given by the persons (ID) .
For example : If a person answered 'No' to the question 1 'Do you have kids' , 'No' to question 2 'Have you ever been married', and 'NO' to the question 3' Did you went to college' the value asigned to the column 'TEST' should be 'A' . But if he ansewerd 'No' to question 1 ,'Yes'to question 2 and 'Yes' to question 3 the value asigned to column test should be 'B'.
I was hoping to find a solution to my problem using the dplyr package for creating a new column 'TEST' :
Here is my code, but it do not work :
df %>%
group_by(ID) %>%
mutate(TEST = ifelse ( (Question == '1' & Answer == "No") & (Question == '2' & Answer == "No") & (Question == '3' & Answer == "No") , 'A'
ifelse ( (Question == '1' & Answer == "No") & (Question == '2' & Answer == "Yes") & (Question == '3' & Answer == "Yes") , 'B'
ifelse ( (Question == '1' & Answer == "Yes") & (Question == '2' & Answer == "Yes") & (Question == '3' & Answer == "No") , 'C'
ifelse( (Question == '1' & Answer == "Yes") & (Question == '2' & Answer == "No") & (Question == '3' & Answer == "No") , 'D' )
)))
The result of the code I am looking for should have this result in this particular example::
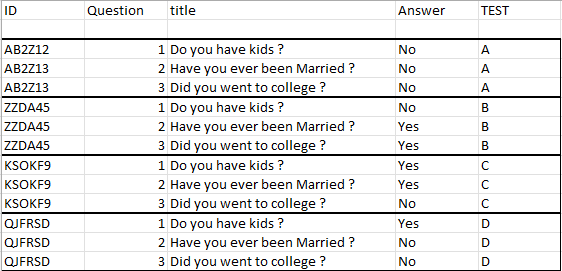
Does anyone have an idea about how to do this in dplyr? This data frame is just an example, the data frames I am dealing with are much larger. Because of its speed I tried to use dplyr, but perhaps there are other, better ways to handle this problem?
CodePudding user response:
case_when() allows for easy chaining of if_else() statements
The way your data is currently structured doesn't cleanly work with the approach you're trying to take so I suggest pivoting it wider (using pivot_wider) to make it easier to work with.
Here's an example of that workflow.
library(tidyverse)
df <- tibble(ID = "ID",
Question = c("Q1","Q2","Q3"),
Answer = c("Yes", "No", "Yes"))
df %>%
pivot_wider(names_from = Question, values_from = Answer) %>%
mutate(TEST = case_when(Q1 == "Yes" & Q2 == "Yes" & Q3 == "Yes" ~ "A",
Q1 == "Yes" & Q2 == "Yes" & Q3 == "No" ~ "B",
Q1 == "Yes" & Q2 == "No" & Q3 == "Yes" ~ "C",
Q1 == "Yes" & Q2 == "No" & Q3 == "No" ~ "D",
Q1 == "No" & Q2 == "Yes" & Q3 == "Yes" ~ "E",
Q1 == "No" & Q2 == "Yes" & Q3 == "No" ~ "F",
Q1 == "No" & Q2 == "No" & Q3 == "Yes" ~ "G",
Q1 == "No" & Q2 == "No" & Q3 == "No" ~ "H"))
# # A tibble: 1 × 5
# ID Q1 Q2 Q3 TEST
# <chr> <chr> <chr> <chr> <chr>
# 1 ID Yes No Yes C
CodePudding user response:
I suggest an alternative method to case_when is to left-join to the pivoted data. While it will effect the same result, it may be easier to read and/or maintain.
ref <- tibble::tribble(
~Q1 , ~Q2 , ~Q3 , ~TEST
, "No" , "No" , "No" , "A"
, "No" , "Yes", "Yes", "B"
, "Yes", "Yes", "No" , "C"
, "Yes", "No" , "No" , "D"
)
Using ref above may be easier to maintain, as it can be edited in any table-editing format (Excel, Calc, text editor, etc). While not difficult, editing a case_when can be prone to a few more typos and/or mistakes.
From here,
out <- df %>%
select(ID, Question, Answer) %>% # we don't need title, or others
tidyr::pivot_wider(names_from = Question, values_from = Answer) %>%
left_join(ref, by = c("Q1", "Q2", "Q3"))
out
# # A tibble: 2 x 5
# ID Q1 Q2 Q3 TEST
# <chr> <chr> <chr> <chr> <chr>
# 1 AB2Z12 No No No A
# 2 ZZDA45 No Yes Yes B
Note that combinations not present (e.g., "No", "Yes", "No") will produce TEST = NA. This can be dealt with in other ways, either by explicitly covering all 8 permutations with TEST values, or by replacing NA later with something else.
To bring this back to the original data, we can join this back on df:
left_join(df, out, by = "ID")
# # A tibble: 6 x 8
# ID Question title Answer Q1 Q2 Q3 TEST
# <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
# 1 AB2Z12 Q1 kids? No No No No A
# 2 AB2Z12 Q2 married? No No No No A
# 3 AB2Z12 Q3 college? No No No No A
# 4 ZZDA45 Q1 kids? No No Yes Yes B
# 5 ZZDA45 Q2 married? Yes No Yes Yes B
# 6 ZZDA45 Q3 college? Yes No Yes Yes B
Data
df <- structure(list(ID = c("AB2Z12", "AB2Z12", "AB2Z12", "ZZDA45", "ZZDA45", "ZZDA45"), Question = c("Q1", "Q2", "Q3", "Q1", "Q2", "Q3"), title = c("kids?", "married?", "college?", "kids?", "married?", "college?"), Answer = c("No", "No", "No", "No", "Yes", "Yes")), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA, -6L))
