I need to plot a HEATMAP in python using x, y, z data from the excel file.
All the values of z are 1 except at (x=5,y=5). The plot should be red at point (5,5) and blue elsewhere. But I am getting false alarms which need to be removed. The COLORMAP I have used is 'jet'
X=[0,0,0,0,0,0,0,0,0,0,1,1,1,1,1,1,1,1,1,1,2,2,2,2,2,2,2,2,2,2,3,3,3,3,3,3,3,3,3,3,4,4,4,4,4,4,4,4,4,4,5,5,5,5,5,5,5,5,5,5,6,6,6,6,6,6,6,6,6,6,7,7,7,7,7,7,7,7,7,7,8,8,8,8,8,8,8,8,8,8,9,9,9,9,9,9,9,9,9,9]
Y=[0,1,2,3,4,5,6,7,8,9,0,1,2,3,4,5,6,7,8,9,0,1,2,3,4,5,6,7,8,9,0,1,2,3,4,5,6,7,8,9,0,1,2,3,4,5,6,7,8,9,0,1,2,3,4,5,6,7,8,9,0,1,2,3,4,5,6,7,8,9,0,1,2,3,4,5,6,7,8,9,0,1,2,3,4,5,6,7,8,9,0,1,2,3,4,5,6,7,8,9]
Z=[1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,9,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1]
Code I have used is:
import matplotlib.pyplot as plt
import numpy as np
from numpy import ravel
from scipy.interpolate import interp2d
import pandas as pd
import matplotlib as mpl
excel_data_df = pd.read_excel('test.xlsx')
X= excel_data_df['x'].tolist()
Y= excel_data_df['y'].tolist()
Z= excel_data_df['z'].tolist()
x_list = np.array(X)
y_list = np.array(Y)
z_list = np.array(Z)
# f will be a function with two arguments (x and y coordinates),
# but those can be array_like structures too, in which case the
# result will be a matrix representing the values in the grid
# specified by those arguments
f = interp2d(x_list,y_list,z_list,kind="linear")
x_coords = np.arange(min(x_list),max(x_list))
y_coords = np.arange(min(y_list),max(y_list))
z= f(x_coords,y_coords)
fig = plt.imshow(z,
extent=[min(x_list),max(x_list),min(y_list),max(y_list)],
origin="lower", interpolation='bicubic', cmap= 'jet', aspect='auto')
# Show the positions of the sample points, just to have some reference
fig.axes.set_autoscale_on(False)
#plt.scatter(x_list,y_list,400, facecolors='none')
plt.xlabel('X Values', fontsize = 15, va="center")
plt.ylabel('Y Values', fontsize = 15,va="center")
plt.title('Heatmap', fontsize = 20)
plt.tight_layout()
plt.show()
For your ease you can also use the X, Y, Z arrays instead of reading excel file.
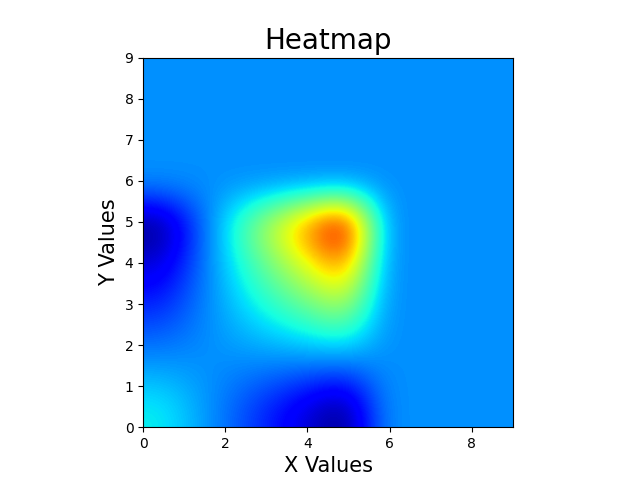
The result that I am getting is:
Here you can see dark blue regions at (5,0) and (0,5). These are the FALSE ALARMS I am getting and I need to REMOVE these.
I am probably doing some beginner's mistake. Grateful to anyone who points it out. Regards
CodePudding user response:
There are at least three problems in your example:
x_coordsandy_coordsare not properly resampled;- the interpolation
zdoes to fill in the whole grid leading to incorrect output; - the output is then forced to be plotted on the original grid (
extent) that add to the confusion.
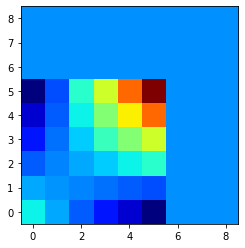
Leading to the following interpolated results:
On what you have applied an extra smoothing with imshow.
Let's create your artificial input:
import matplotlib.pyplot as plt
import numpy as np
x = np.arange(0, 11)
y = np.arange(0, 11)
X, Y = np.meshgrid(x, y)
Z = np.ones(X.shape)
Z[5,5] = 9
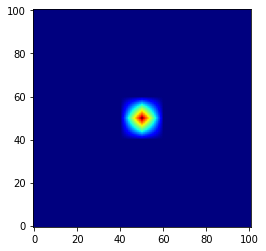
Depending on how you want to proceed, you can simply let imshow smooth your signal by interpolation:
fig, axe = plt.subplots()
axe.imshow(Z, origin="lower", cmap="jet", interpolation='bicubic')
And you are done, simple and efficient!
If you aim to do it by yourself, then choose the interpolant that suits you best and resample on a grid with a higher resolution:
interpolant = interpolate.interp2d(x, y, Z.ravel(), kind="linear")
xlin = np.linspace(0, 10, 101)
ylin = np.linspace(0, 10, 101)
zhat = interpolant(xlin, ylin)
fig, axe = plt.subplots()
axe.imshow(zhat, origin="lower", cmap="jet")