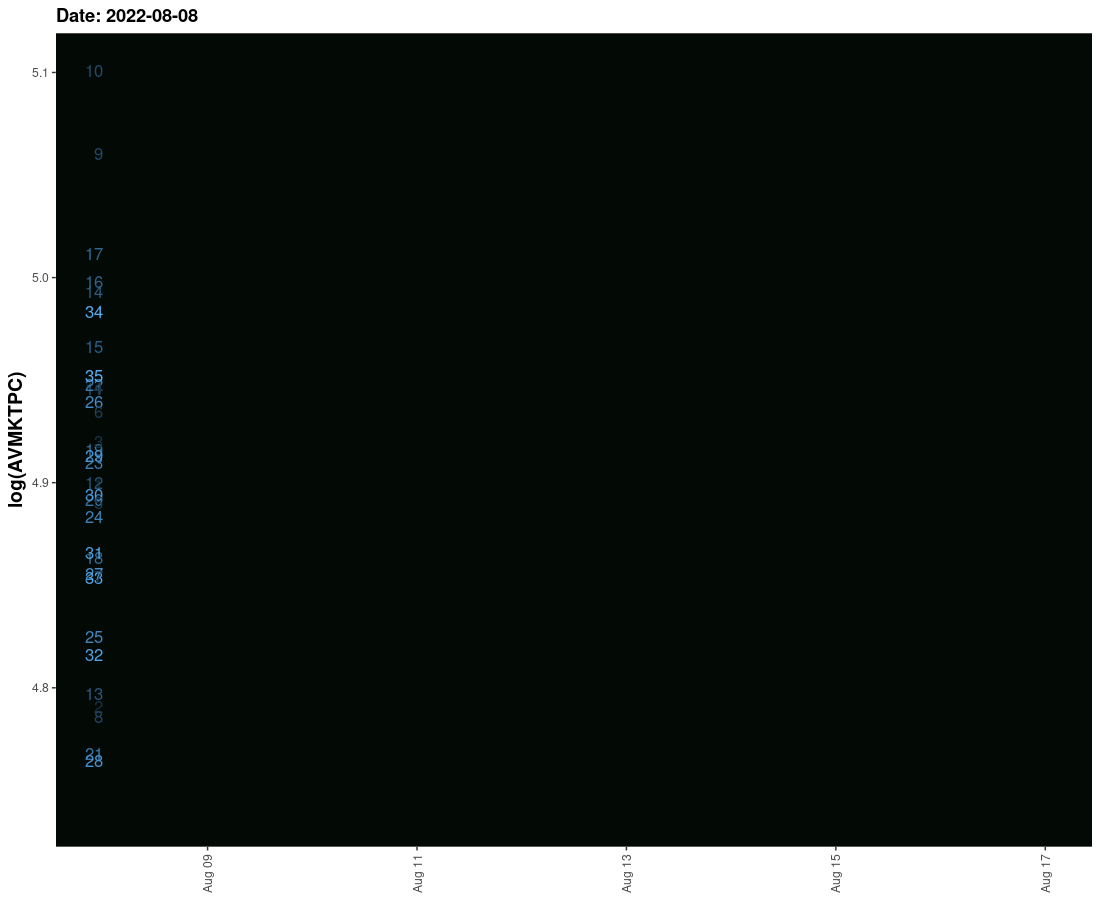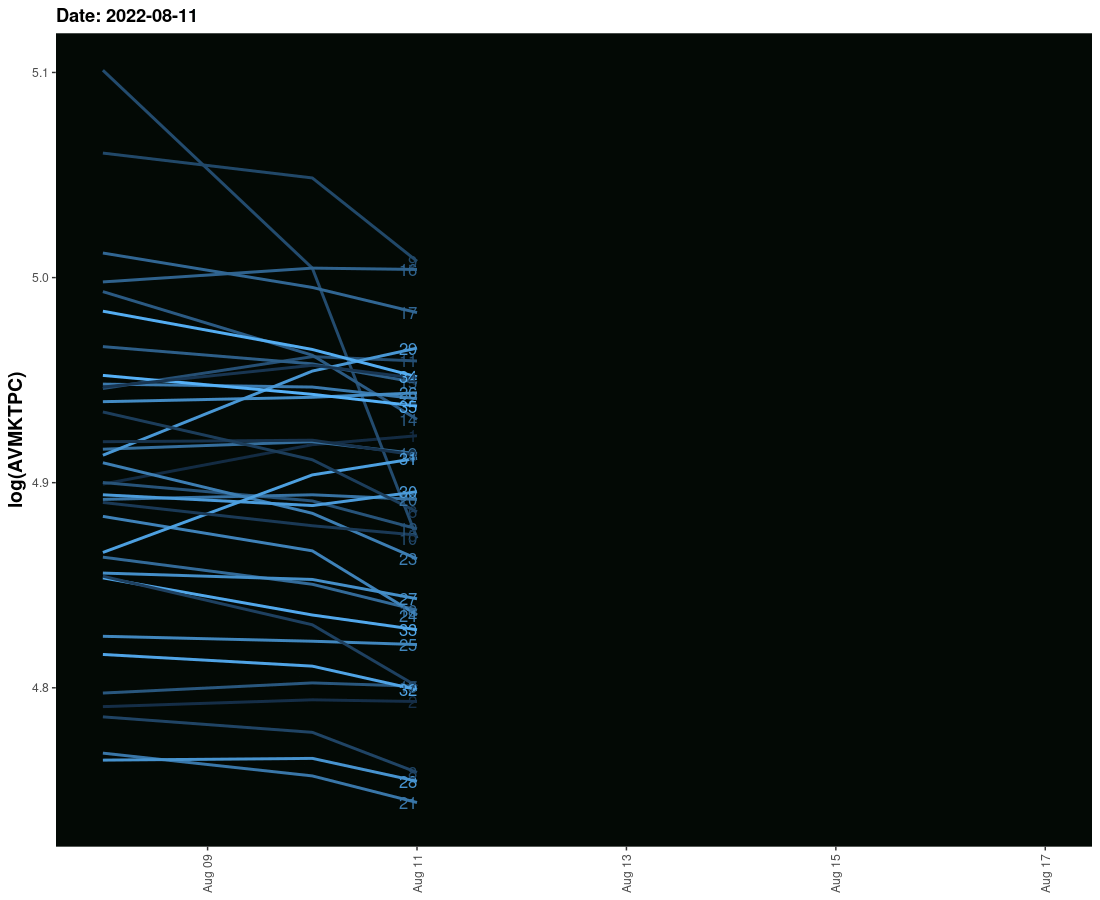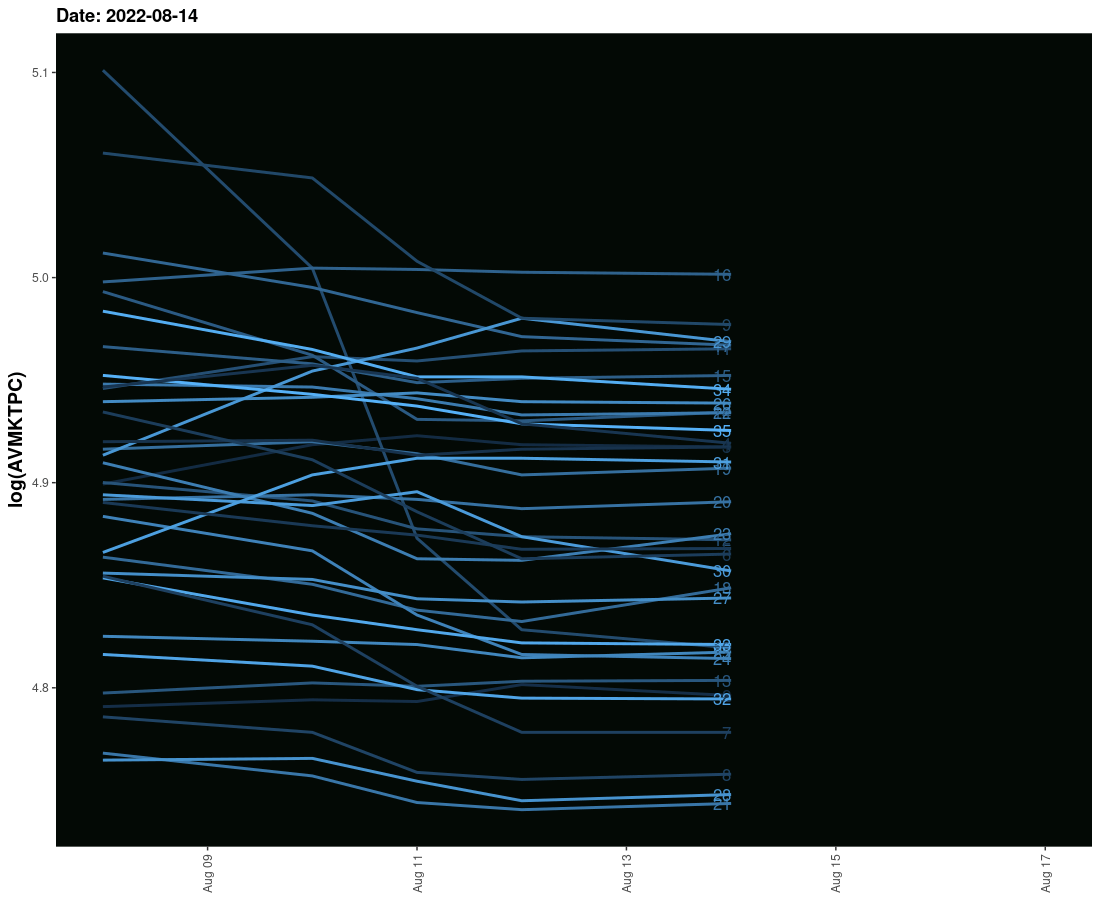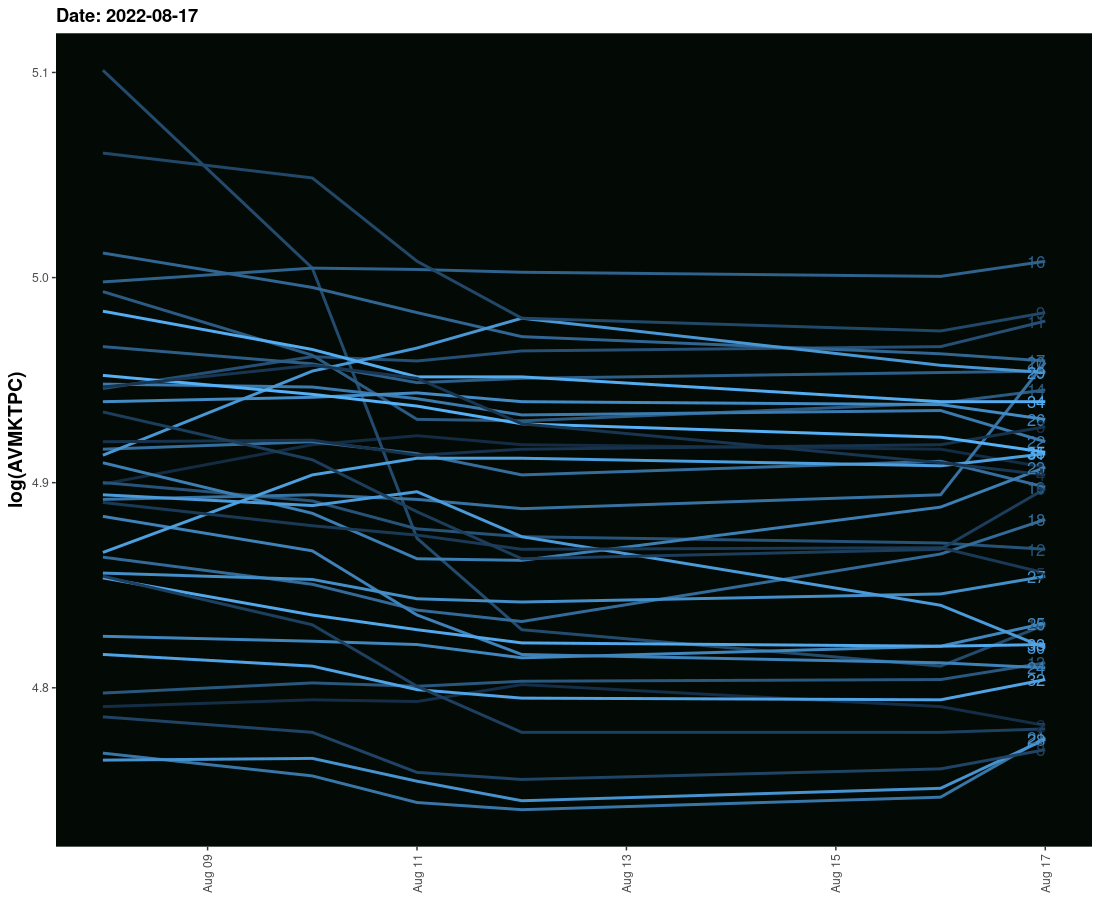
I have tried to create an animated graph for which I am using geom_smooth that moves and also the labels that move with the lines. However, I am not able to provide space between the line and labels so their legibility is affected. Need to solve this.
The code for plotting the animation is as follows.
p <- ggplot(tbl66, aes(x=TRDAY, y=log(AVMKTPC), col=LABEL2, group = LABEL2)) geom_line(size=1)
theme(legend.position="none")
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
geom_dl(aes(label = LABEL2), method = list(dl.combine("first.points")), cex = 1)
theme(panel.background = element_rect(fill = "#030905"))
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
transition_reveal(TRDAY)
ease_aes('cubic-in-out')
labs(title = 'Date: {frame_along}')
theme(legend.position = 'none',
title = element_text(size = 11, face = "bold"),
axis.title.x = element_blank(),
axis.title = element_text(size = 14, face = "bold"),
strip.text = element_text(size = 14, face = "bold"),
plot.caption = element_text(face = "italic"))
animate(p, 10, .2, height=900, width=1100, res=100)
anim_save("codovid.gif")
The sample data is given here. It provides three columns one is the date column and the second is the value column and the third is the label column. I need these values to run ahead of line with some space.
structure(list(TRDAY = structure(c(1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600, 1659897000, 1660069800, 1660156200,
1660242600, 1660588200, 1660674600), class = c("POSIXct", "POSIXt"
), tzone = ""), AVMKTPC = c(134.2, 136.8, 137.4, 136.8, 136.5,
135.3, 120.4, 120.8, 120.7, 121.7, 120.4, 119.3, 137, 137.1,
136.1, 136.5, 136.8, 138, 140.7, 142.2, 141.3, 138.2, 135.6,
134.8, 133, 131.5, 130.9, 130, 130.1, 128.5, 139, 135.8, 132.4,
129.4, 130, 133.9, 128.3, 125.3, 121.6, 118.9, 118.9, 119.1,
119.8, 118.9, 116.6, 116.2, 116.8, 117.9, 157.7, 155.8, 149.6,
145.5, 144.6, 145.9, 164.2, 149.1, 130.7, 125, 122.8, 125.3,
140.6, 142.8, 142.5, 143.2, 143.5, 145.3, 134.3, 133.1, 131.3,
130.8, 130.4, 130, 121.2, 121.8, 121.6, 121.9, 122, 123, 147.4,
142.9, 138.5, 138.4, 139.6, 140.5, 143.5, 142.3, 141, 141.3,
141.7, 141.8, 148.1, 149.1, 149, 148.8, 148.5, 149.6, 150.2,
147.7, 145.9, 144.2, 143, 142.5, 129.5, 127.8, 126.2, 125.5,
129.7, 131.9, 136.5, 137, 136.2, 134.8, 135.7, 134, 133.2, 133.5,
133.2, 132.6, 133.5, 142.4, 117.7, 116.4, 114.9, 114.5, 115.2,
118.6, 140.9, 140.7, 139.9, 138.8, 139.1, 137, 135.6, 132.3,
129.4, 129.3, 132.7, 135.3, 132.1, 129.9, 125.9, 123.5, 123,
122.7, 124.6, 124.3, 124.1, 123.3, 124, 125.4, 139.7, 140, 140.3,
139.7, 139.5, 138.5, 128.5, 128.1, 126.9, 126.7, 127.2, 128.3,
117.3, 117.4, 116.1, 115, 115.7, 118.5, 136.1, 141.8, 143.4,
145.5, 142.2, 141.7, 133.5, 132.8, 133.7, 130.8, 126.5, 123.9,
129.8, 134.8, 135.9, 135.9, 135.4, 136.2, 123.5, 122.8, 121.4,
120.9, 120.8, 122, 128.2, 125.9, 125, 124.2, 124, 124.1, 146,
143.3, 141.4, 141.4, 139.7, 139.7, 141.5, 140.2, 139.4, 138.2,
137.3, 136.3), LABEL2 = c(1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L,
2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 5L,
5L, 5L, 5L, 5L, 5L, 6L, 6L, 6L, 6L, 6L, 6L, 7L, 7L, 7L, 7L, 7L,
7L, 8L, 8L, 8L, 8L, 8L, 8L, 9L, 9L, 9L, 9L, 9L, 9L, 10L, 10L,
10L, 10L, 10L, 10L, 11L, 11L, 11L, 11L, 11L, 11L, 12L, 12L, 12L,
12L, 12L, 12L, 13L, 13L, 13L, 13L, 13L, 13L, 14L, 14L, 14L, 14L,
14L, 14L, 15L, 15L, 15L, 15L, 15L, 15L, 16L, 16L, 16L, 16L, 16L,
16L, 17L, 17L, 17L, 17L, 17L, 17L, 18L, 18L, 18L, 18L, 18L, 18L,
19L, 19L, 19L, 19L, 19L, 19L, 20L, 20L, 20L, 20L, 20L, 20L, 21L,
21L, 21L, 21L, 21L, 21L, 22L, 22L, 22L, 22L, 22L, 22L, 23L, 23L,
23L, 23L, 23L, 23L, 24L, 24L, 24L, 24L, 24L, 24L, 25L, 25L, 25L,
25L, 25L, 25L, 26L, 26L, 26L, 26L, 26L, 26L, 27L, 27L, 27L, 27L,
27L, 27L, 28L, 28L, 28L, 28L, 28L, 28L, 29L, 29L, 29L, 29L, 29L,
29L, 30L, 30L, 30L, 30L, 30L, 30L, 31L, 31L, 31L, 31L, 31L, 31L,
32L, 32L, 32L, 32L, 32L, 32L, 33L, 33L, 33L, 33L, 33L, 33L, 34L,
34L, 34L, 34L, 34L, 34L, 35L, 35L, 35L, 35L, 35L, 35L)), row.names = c(NA,
-210L), class = c("tbl_df", "tbl", "data.frame"))
CodePudding user response:
One option would be to use geom_text(aes(label = LABEL2), hjust = -0.1) to left align the labels with the data points and to add some padding:
library(gganimate)
#> Loading required package: ggplot2
p <- ggplot(tbl66, aes(x=TRDAY, y=log(AVMKTPC), col=LABEL2, group = LABEL2)) geom_line(size=1)
theme(legend.position="none")
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
geom_text(aes(label = LABEL2), hjust = -0.1)
theme(panel.background = element_rect(fill = "#030905"))
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
transition_reveal(TRDAY)
ease_aes('cubic-in-out')
labs(title = 'Date: {frame_along}')
theme(legend.position = 'none',
title = element_text(size = 11, face = "bold"),
axis.title.x = element_blank(),
axis.title = element_text(size = 14, face = "bold"),
strip.text = element_text(size = 14, face = "bold"),
plot.caption = element_text(face = "italic"))
animate(p, 10, .2, height=900, width=1100, res=100)
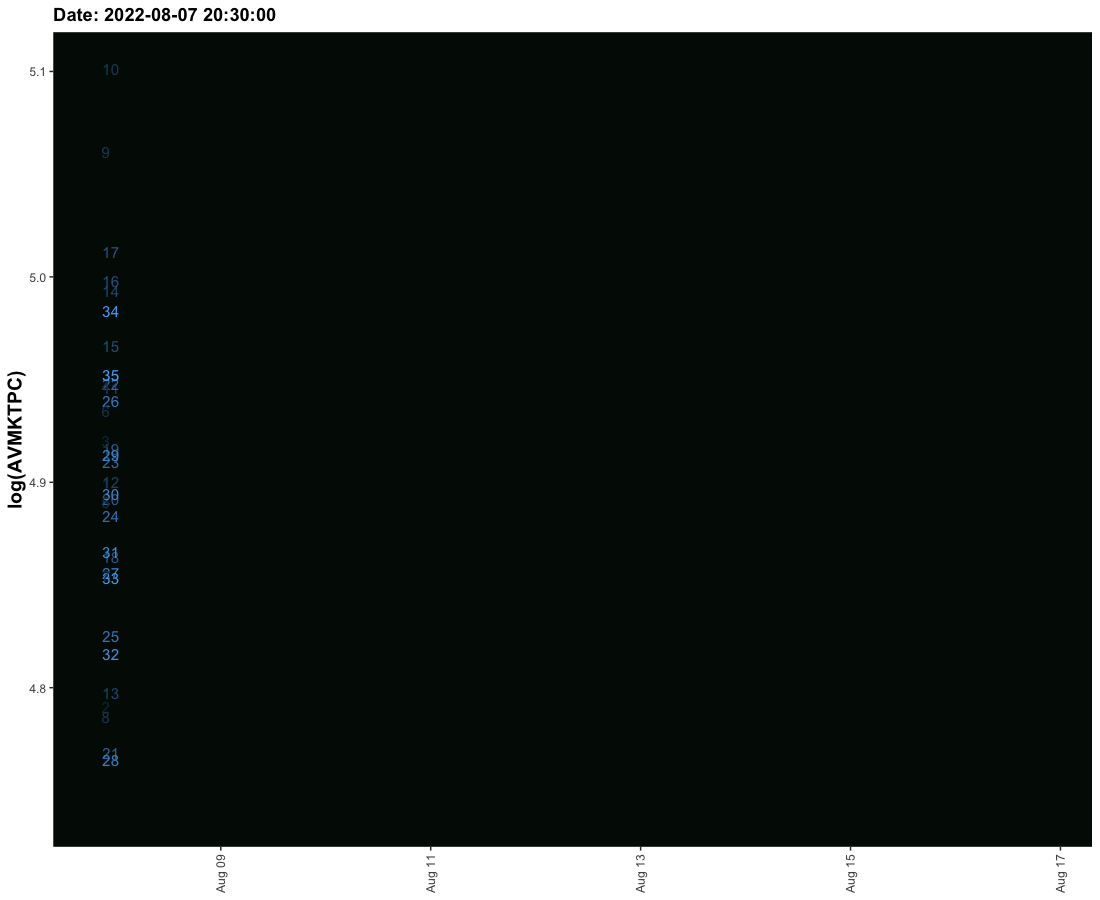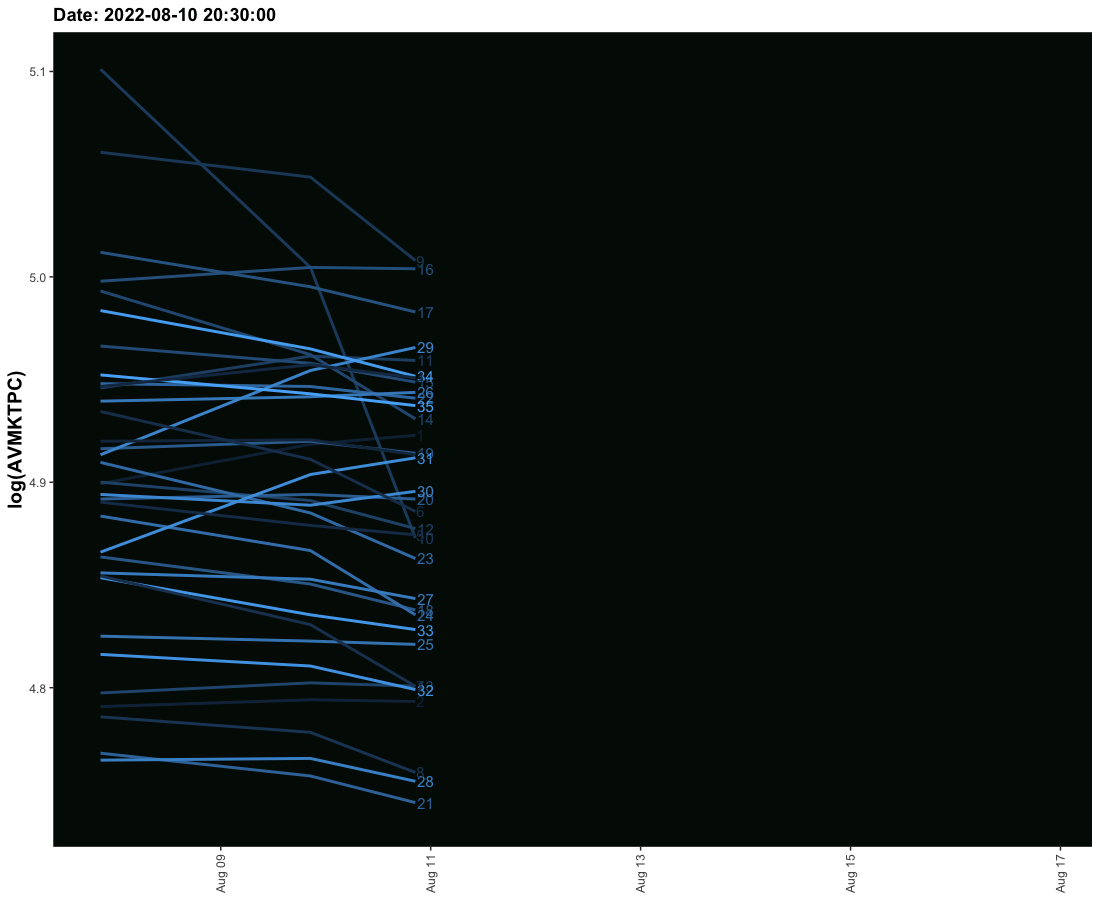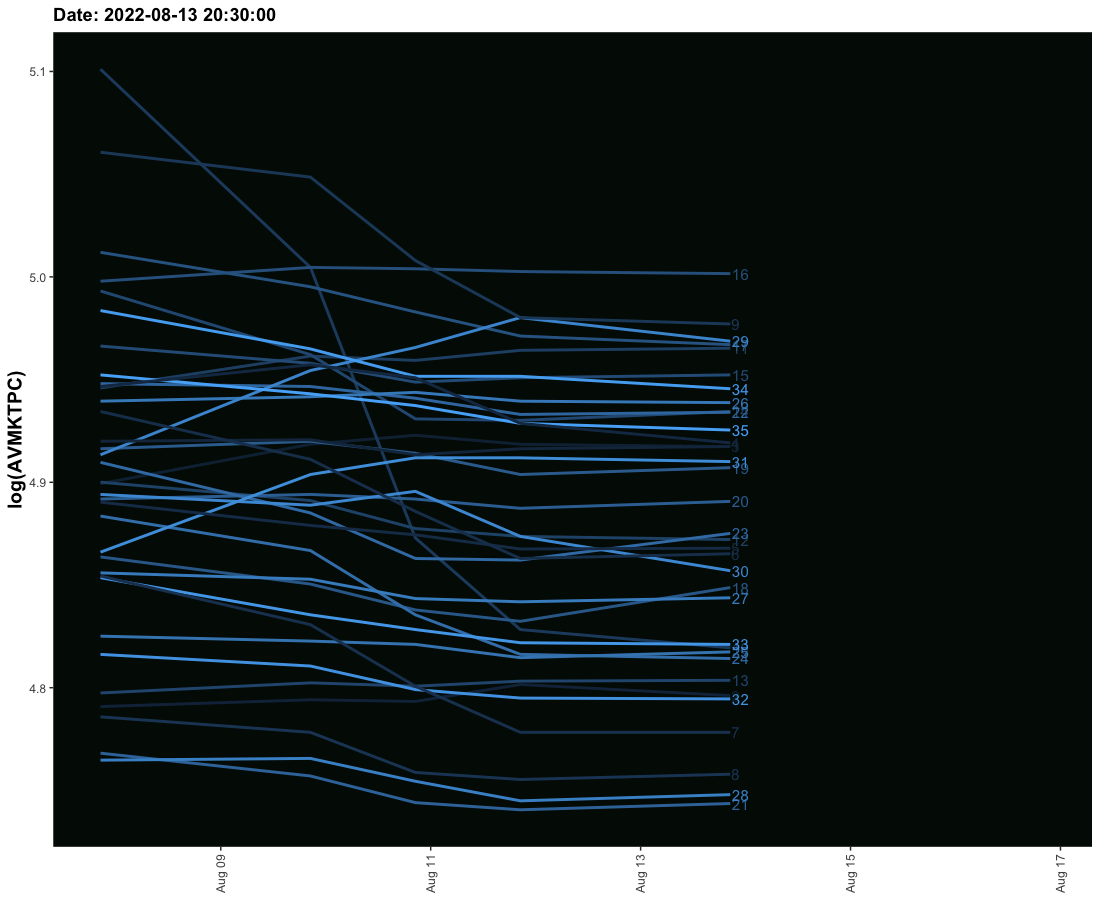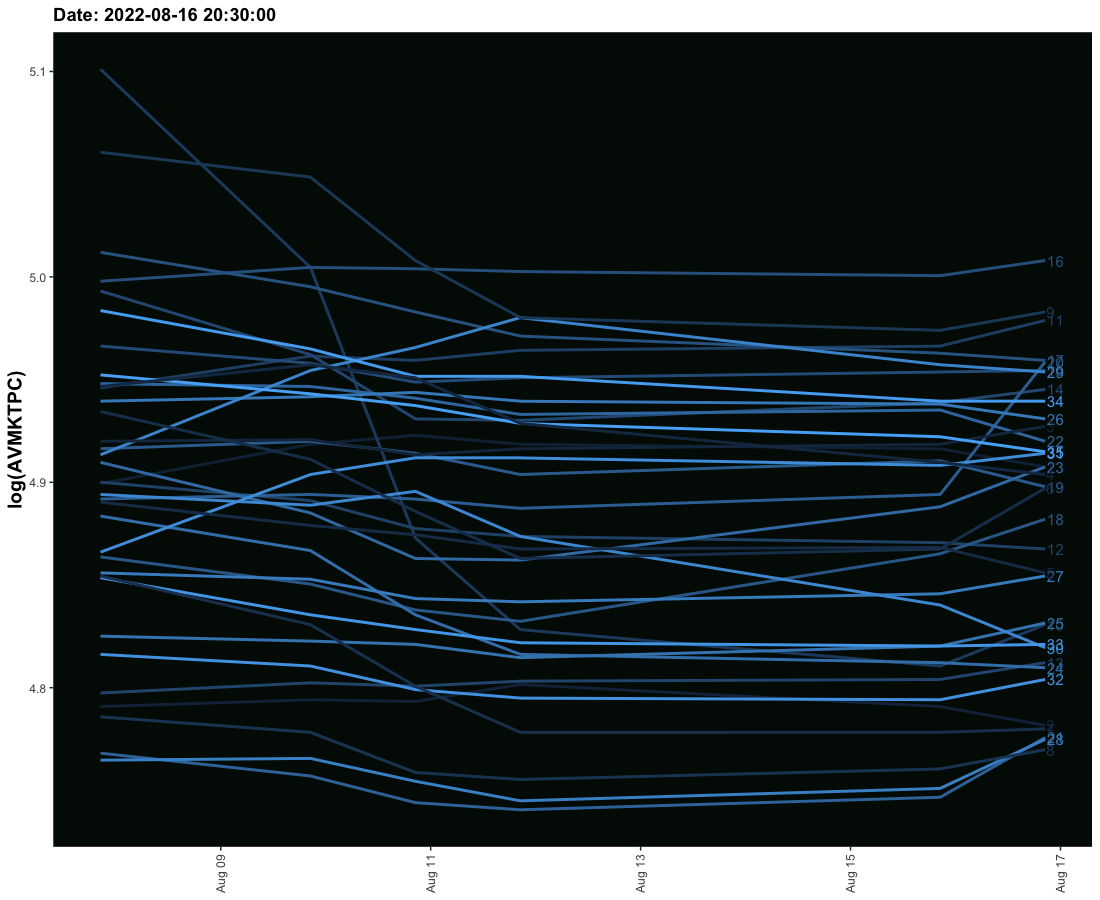
# anim_save("codovid.gif")