I have a dataset like the following where one customer can belong to multiple departments
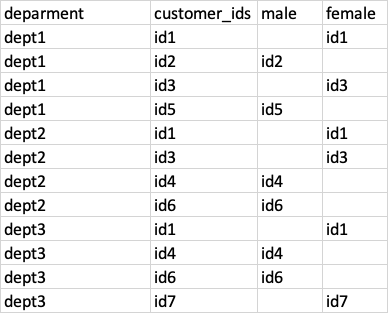
Now I want to aggregate the number of customers exist in either of the 2 departments. For example see the below result
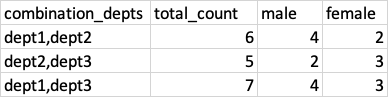
The first aggregation example
dept1 = {id1, id2, id3, id5}
dept2 = {id1, id3, id4, id6}
dept1 Union dept2 => dept1,dept2 = {id1, id2, id3, id5} U {id1, id3, id4, id6}
dept1,dept2 = {id1, id2, id3, id5, id4, id6}
dept1,dept2 = 6
Similarly, female and male Ids are aggregated to 4 and 2. I'm new to Pyspark so I'm not sure how to do this kind of aggregation with Pyspark. Any hint or example solution would be great.
CodePudding user response:
There is probably a simpler solution but I thought about using PySpark arrays.
Steps:
- agg all employoees to each department
- cross join the df to itself to get all combinations of departments, of course filter to get half of them. then UNION the two sets and EXPLODE them to get back to a "normal" dataframe without arrays.
Note: probably won't work if you have departments with 100k employees.
df_agg = df \
.groupBy('department') \
.agg(collect_set('customer_ids').alias('customer_ids_array'))
distinct_employees = df_agg.alias('df1') \
.crossJoin(df_agg.alias('df2')) \
.filter(col('df1.department') < col('df2.department')) \
.select(
concat('df1.department', lit(','), 'df2.department').alias('combinations_depts'),
explode(array_union('df1.department', 'df2.department')).alias('employee')
)
from here join to the orig df to get the statistics you want with normal count function.
CodePudding user response:
You could create arrays of distinct ids and use size() to count the number of ids. Arrays of unique ids can be created using collect_set. Here's an example.
Given an input data (say, data_sdf) as follows.
# ----- --- ---- ----
# | dept| id| f| m|
# ----- --- ---- ----
# |dept1|id3| id3|null|
# |dept3|id3| id3|null|
# |dept2|id5|null| id5|
# |dept2|id1| id1|null|
# |dept1|id2|null| id2|
# |dept3|id1| id1|null|
# |dept2|id2|null| id2|
# |dept1|id1| id1|null|
# |dept3|id2|null| id2|
# ----- --- ---- ----
Create arrays of distinct ids using collect_set for all departments.
data_sdf. \
groupBy('dept'). \
agg(func.collect_set('id').alias('unique_ids'),
func.collect_set('f').alias('unique_f_ids'),
func.collect_set('m').alias('unique_m_ids')
). \
show()
# ----- --------------- ------------ ------------
# | dept| unique_ids|unique_f_ids|unique_m_ids|
# ----- --------------- ------------ ------------
# |dept3|[id2, id1, id3]| [id1, id3]| [id2]|
# |dept1|[id2, id1, id3]| [id1, id3]| [id2]|
# |dept2|[id2, id5, id1]| [id1]| [id2, id5]|
# ----- --------------- ------------ ------------
Now, use window function to create combination of consecutive 2 departments and their unique ids and use size to count the number of ids in each array. The idea is to have a sorting column that would tell spark the departments' sequence. For this example, I've use the dept field itself as it had an integer identifier.
data_sdf. \
groupBy('dept'). \
agg(func.collect_set('id').alias('unique_ids'),
func.collect_set('f').alias('unique_f_ids'),
func.collect_set('m').alias('unique_m_ids')
). \
withColumn('comb_dept',
func.concat_ws(',',func.collect_list('dept').over(wd.orderBy('dept').rowsBetween(-1, 0)))
). \
withColumn('comb_unq_ids',
func.array_distinct(func.flatten(func.collect_list('unique_ids').over(wd.orderBy('dept').rowsBetween(-1, 0))))
). \
withColumn('comb_unq_f_ids',
func.array_distinct(func.flatten(func.collect_list('unique_f_ids').over(wd.orderBy('dept').rowsBetween(-1, 0))))
). \
withColumn('comb_unq_m_ids',
func.array_distinct(func.flatten(func.collect_list('unique_m_ids').over(wd.orderBy('dept').rowsBetween(-1, 0))))
). \
filter(func.col('dept') != func.col('comb_dept')). \
selectExpr('comb_dept', 'size(comb_unq_ids) as tot_count', 'size(comb_unq_f_ids) as f_count', 'size(comb_unq_m_ids) as m_count'). \
show()
# ----------- --------- ------- -------
# | comb_dept|tot_count|f_count|m_count|
# ----------- --------- ------- -------
# |dept1,dept2| 4| 2| 2|
# |dept2,dept3| 4| 2| 2|
# ----------- --------- ------- -------
