I know there have been several topics about this question already, but non of the answers solved my problem. I'm trying to run a random forest model, using caret. I'm getting this error:
Something is wrong; all the ROC metric values are missing:
ROC Sens Spec
Min. : NA Min. : NA Min. : NA
1st Qu.: NA 1st Qu.: NA 1st Qu.: NA
Median : NA Median : NA Median : NA
Mean :NaN Mean :NaN Mean :NaN
3rd Qu.: NA 3rd Qu.: NA 3rd Qu.: NA
Max. : NA Max. : NA Max. : NA
NA's :10 NA's :10 NA's :10
Error: Stopping
In addition: There were 50 or more warnings (use warnings() to see the first 50)
What is this error and how do I fix it? I should note that the data I provide here is abbreviated, my data is much bigger with more predictive features, but I got the same error there also. Therefore I tried to make the model based on simpler data.
I have no NA or Inf values in the data. I tried redownloading packages like caret and pROC, I tried to change the parameters, but nothing helped.
This is the code:
ctrlCV = trainControl(method = 'cv', number = 10 , classProbs = TRUE , savePredictions = TRUE, summaryFunction = twoClassSummary )
rfGRID <- expand.grid(.mtry = c(1 : 10))
rfFit <- train(response~., data = dimdum,
method = "rf",
metric="ROC",
importance = TRUE,
trControl = ctrlCV,
tuneGrid = rfGRID,
ntree = c(50,100,150,200,300,400,500),
nodesize = c(1:10)
)
rfROC = roc(dimdum$response,predict(rfFit,dimdum, type='prob')[,1])
plot(rfROC)
Some of the data:
structure(list(response = c("NoResponse", "Response", "NoResponse",
"NoResponse", "NoResponse", "Response", "NoResponse", "NoResponse",
"NoResponse", "NoResponse", "NoResponse", "Response", "NoResponse",
"Response", "NoResponse", "NoResponse", "NoResponse", "NoResponse",
"Response", "Response", "NoResponse", "NoResponse", "NoResponse",
"NoResponse", "NoResponse", "NoResponse", "NoResponse", "Response",
"NoResponse", "NoResponse", "NoResponse", "NoResponse", "NoResponse",
"NoResponse", "NoResponse", "Response", "NoResponse", "NoResponse",
"NoResponse", "Response", "Response", "NoResponse", "Response",
"Response", "NoResponse", "Response", "Response", "NoResponse",
"Response", "Response"), CD4..Tem = c(0.206146305909711, 0.38344530718027,
0.111171710498514, -0.024620418652091, -0.024620418652091, 0.241356282324198,
0.190401963339481, 0.0381564797828905, 0.00208479038732372, -0.024620418652091,
0.0345146685774692, 0.0047448955916752, 0.0954402244646442, 0.216170609750478,
0.0679837609588422, 0.1087338604344, 0.0307783462567513, -0.024620418652091,
0.00930882669937516, 0.228984175232275, 0.198029266287967, 0.00849741399216577,
0.167122425878708, -0.0185389752646852, 0.0349285293854749, -0.015164453751509,
0.0530002007752186, -0.00464766527016771, 0.229228539194469,
-0.024620418652091, -0.024620418652091, 0.00909241866793368,
0.00959645779130966, -0.0169517988930254, 0.130416251320013,
-0.0226155780862924, -0.0226155780862924, -0.00708354014661853,
-0.0226155780862924, 0.0236200668251617, -0.0226155780862924,
-0.0226155780862924, -0.0226155780862924, -0.0226155780862924,
-0.0226155780862924, -0.00226780446329141, -0.00703727203694584,
-0.0226155780862924, 0.040447933249888, -0.0226155780862924),
Epithelial.cells = c(0.213818759771441, 0.224884228557244,
0.213818759771441, 0.564636116181376, 0.213818759771441,
0.213818759771441, 0.256571259511661, 0.213818759771441,
0.225551386999972, 0.213818759771441, 0.320498217450289,
0.213818759771441, 0.213818759771441, 0.213818759771441,
0.250752952186148, 0.54432086478806, 0.213818759771441, 0.213818759771441,
0.213818759771441, 0.224579338204213, 0.244604368723937,
0.239048638424405, 0.213818759771441, 0.213818759771441,
0.213818759771441, 0.213818759771441, 0.755246080444261,
0.213818759771441, 0.224789200187943, 0.213818759771441,
0.213818759771441, 0.213818759771441, 0.602551670320415,
0.221211285726714, 0.528154858032774, 0.191393513022707,
0.204473730554233, 0.199140565064947, 0.191393513022707,
0.191393513022707, 0.208364515830724, 0.199044034497245,
0.218231966624601, 0.191393513022707, 0.199037583564646,
0.233310726880044, 0.257245920265987, 0.2245306029313, 0.30356359401388,
0.202283902795669)), row.names = c("Pt1", "Pt101", "Pt106",
"Pt11", "Pt17", "Pt18", "Pt24", "Pt26", "Pt27", "Pt28", "Pt29",
"Pt3", "Pt31", "Pt34", "Pt36", "Pt37", "Pt38", "Pt39", "Pt44",
"Pt49", "Pt5", "Pt52", "Pt59", "Pt62", "Pt65", "Pt66", "Pt67",
"Pt72", "Pt77", "Pt78", "Pt84", "Pt85", "Pt89", "Pt9", "Pt90",
"EA595454", "EA595500", "EA595522", "EA595529", "EA595597", "EA595624",
"EA595635", "EA595647", "EA595654", "EA595719", "EA595720", "EA632133",
"EA632171", "EA632174", "EA632234"), class = "data.frame")
CodePudding user response:
If you test the ntree values one-at-a-time in a loop it works as expected:
library(randomForest)
#> randomForest 4.7-1.1
#> Type rfNews() to see new features/changes/bug fixes.
#library(mlbench)
library(caret)
#> Loading required package: ggplot2
#>
#> Attaching package: 'ggplot2'
#> The following object is masked from 'package:randomForest':
#>
#> margin
#> Loading required package: lattice
library(pROC)
#> Type 'citation("pROC")' for a citation.
#>
#> Attaching package: 'pROC'
#> The following objects are masked from 'package:stats':
#>
#> cov, smooth, var
df <- structure(list(response = c("NoResponse", "Response", "NoResponse",
"NoResponse", "NoResponse", "Response", "NoResponse", "NoResponse",
"NoResponse", "NoResponse", "NoResponse", "Response", "NoResponse",
"Response", "NoResponse", "NoResponse", "NoResponse", "NoResponse",
"Response", "Response", "NoResponse", "NoResponse", "NoResponse",
"NoResponse", "NoResponse", "NoResponse", "NoResponse", "Response",
"NoResponse", "NoResponse", "NoResponse", "NoResponse", "NoResponse",
"NoResponse", "NoResponse", "Response", "NoResponse", "NoResponse",
"NoResponse", "Response", "Response", "NoResponse", "Response",
"Response", "NoResponse", "Response", "Response", "NoResponse",
"Response", "Response"),
CD4..Tem = c(0.206146305909711, 0.38344530718027,
0.111171710498514, -0.024620418652091, -0.024620418652091, 0.241356282324198,
0.190401963339481, 0.0381564797828905, 0.00208479038732372, -0.024620418652091,
0.0345146685774692, 0.0047448955916752, 0.0954402244646442, 0.216170609750478,
0.0679837609588422, 0.1087338604344, 0.0307783462567513, -0.024620418652091,
0.00930882669937516, 0.228984175232275, 0.198029266287967, 0.00849741399216577,
0.167122425878708, -0.0185389752646852, 0.0349285293854749, -0.015164453751509,
0.0530002007752186, -0.00464766527016771, 0.229228539194469,
-0.024620418652091, -0.024620418652091, 0.00909241866793368,
0.00959645779130966, -0.0169517988930254, 0.130416251320013,
-0.0226155780862924, -0.0226155780862924, -0.00708354014661853,
-0.0226155780862924, 0.0236200668251617, -0.0226155780862924,
-0.0226155780862924, -0.0226155780862924, -0.0226155780862924,
-0.0226155780862924, -0.00226780446329141, -0.00703727203694584,
-0.0226155780862924, 0.040447933249888, -0.0226155780862924),
Epithelial.cells = c(0.213818759771441, 0.224884228557244,
0.213818759771441, 0.564636116181376, 0.213818759771441,
0.213818759771441, 0.256571259511661, 0.213818759771441,
0.225551386999972, 0.213818759771441, 0.320498217450289,
0.213818759771441, 0.213818759771441, 0.213818759771441,
0.250752952186148, 0.54432086478806, 0.213818759771441, 0.213818759771441,
0.213818759771441, 0.224579338204213, 0.244604368723937,
0.239048638424405, 0.213818759771441, 0.213818759771441,
0.213818759771441, 0.213818759771441, 0.755246080444261,
0.213818759771441, 0.224789200187943, 0.213818759771441,
0.213818759771441, 0.213818759771441, 0.602551670320415,
0.221211285726714, 0.528154858032774, 0.191393513022707,
0.204473730554233, 0.199140565064947, 0.191393513022707,
0.191393513022707, 0.208364515830724, 0.199044034497245,
0.218231966624601, 0.191393513022707, 0.199037583564646,
0.233310726880044, 0.257245920265987, 0.2245306029313, 0.30356359401388,
0.202283902795669)),
row.names = c("Pt1", "Pt101", "Pt106",
"Pt11", "Pt17", "Pt18", "Pt24", "Pt26", "Pt27", "Pt28", "Pt29",
"Pt3", "Pt31", "Pt34", "Pt36", "Pt37", "Pt38", "Pt39", "Pt44",
"Pt49", "Pt5", "Pt52", "Pt59", "Pt62", "Pt65", "Pt66", "Pt67",
"Pt72", "Pt77", "Pt78", "Pt84", "Pt85", "Pt89", "Pt9", "Pt90",
"EA595454", "EA595500", "EA595522", "EA595529", "EA595597", "EA595624",
"EA595635", "EA595647", "EA595654", "EA595719", "EA595720", "EA632133",
"EA632171", "EA632174", "EA632234"), class = "data.frame")
ctrlCV = trainControl(method = 'cv', number = 10 , classProbs = TRUE , savePredictions = TRUE, summaryFunction = twoClassSummary)
rfGRID <- expand.grid(.mtry = sqrt(ncol(df[-c(1)])))
rfFit <- train(response ~ ., data = df,
method = "rf",
metric = "ROC",
importance = TRUE,
trControl = ctrlCV,
tuneGrid = rfGRID,
ntree = 10,
nodesize = c(1:10)
)
print(rfFit)
#> Random Forest
#>
#> 50 samples
#> 2 predictor
#> 2 classes: 'NoResponse', 'Response'
#>
#> No pre-processing
#> Resampling: Cross-Validated (10 fold)
#> Summary of sample sizes: 45, 45, 44, 46, 45, 44, ...
#> Resampling results:
#>
#> ROC Sens Spec
#> 0.6104167 0.825 0.35
#>
#> Tuning parameter 'mtry' was held constant at a value of 1.414214
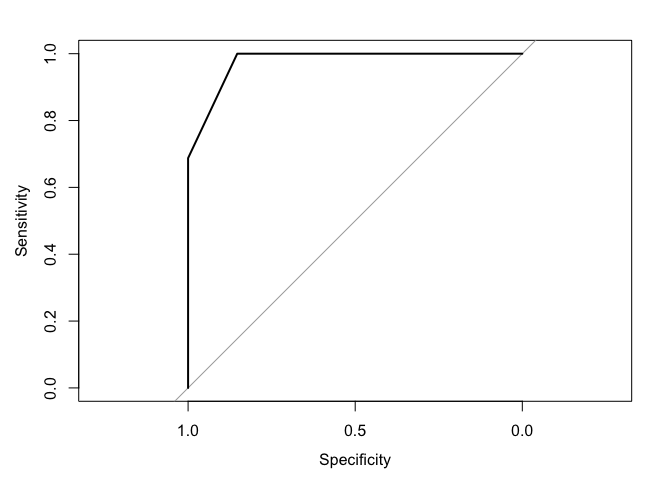
rfROC = roc(df$response, predict(rfFit,df, type='prob')[,1])
#> Setting levels: control = NoResponse, case = Response
#> Setting direction: controls > cases
plot(rfROC)
# Use smaller ntree values for this example
modellist <- list()
for (ntree in c(10,20,50,75,100)){
set.seed(123)
fit <- train(response ~ ., data = df,
method = 'rf',
metric = 'ROC',
importance = TRUE,
trControl = ctrlCV,
tuneGrid = rfGRID,
ntree = ntree)
key <- toString(ntree)
modellist[[key]] <- fit
}
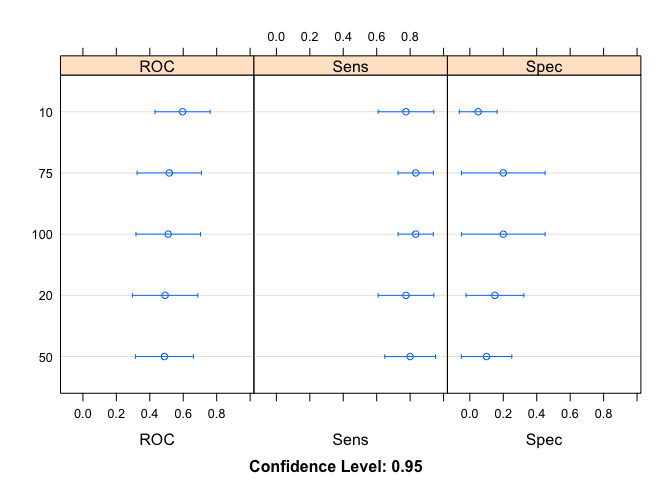
results <- resamples(modellist)
summary(results)
#>
#> Call:
#> summary.resamples(object = results)
#>
#> Models: 10, 20, 50, 75, 100
#> Number of resamples: 10
#>
#> ROC
#> Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
#> 10 0.1666667 0.5208333 0.6458333 0.5958333 0.7500000 0.8333333 0
#> 20 0.0000000 0.2916667 0.5000000 0.4916667 0.6666667 0.9166667 0
#> 50 0.0000000 0.3437500 0.5833333 0.4875000 0.6666667 0.7500000 0
#> 75 0.0000000 0.3750000 0.5000000 0.5166667 0.7291667 0.8333333 0
#> 100 0.0000000 0.3593750 0.5000000 0.5104167 0.7291667 0.8333333 0
#>
#> Sens
#> Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
#> 10 0.3333333 0.6875 0.75 0.7750000 1 1 0
#> 20 0.3333333 0.6875 0.75 0.7750000 1 1 0
#> 50 0.3333333 0.7500 0.75 0.8000000 1 1 0
#> 75 0.6666667 0.7500 0.75 0.8333333 1 1 0
#> 100 0.6666667 0.7500 0.75 0.8333333 1 1 0
#>
#> Spec
#> Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
#> 10 0 0 0 0.05 0.000 0.5 0
#> 20 0 0 0 0.15 0.375 0.5 0
#> 50 0 0 0 0.10 0.000 0.5 0
#> 75 0 0 0 0.20 0.375 1.0 0
#> 100 0 0 0 0.20 0.375 1.0 0
dotplot(results)
Created on 2022-09-02 by the reprex package (v2.0.1)
