I have one data below
df<-structure(list(SEASON = c("Low D", "Low D", "Low D", "Low D", "Low D", "Low D", "Low D", "Low D", "Low D", "Low D", "High D", "High D", "High D", "High D", "High D", "High D", "High D", "High D", "High D", "High D"), GENOTYPE = c("A", "B", "C", "D", "E", "F", "G", "H", "I", "J", "A", "B", "C", "D", "E", "F", "G", "H", "I", "J"), Y1 = c(16.95, 16.25, 16.1, 16.25, 17.25, 17.7, 15.75, 17.5, 16.9, 16.95, 15.95, 16.15, 15.4, 15.45, 16.5, 17, 15.8, 17.5, 16.55, 16.95), Y2 = c(36.325, 35.425, 38.175, 36.3, 35.225, 36, 35, 33.15, 34.975, 35.35, 29.55, 30.225, 33.35, 30.7, 30.35, 30.8, 31.95, 27.5, 30.075, 28.825), Y3 = c(242.475, 237.15, 234.307692307692, 243.75, 232.575, 257.4, 223.875, 241.35, 235.65, 239.275, 163.85, 177.4, 158.675, 171.95, 161.625, 181.65, 173.175, 166.05, 174.717948717949, 178.875), Y4 = c(206.375, 206.525, 206.230769230769, 210.925, 201.7, 223.5, 199.725, 211.8, 206.525, 211.75, 141.175, 154.075, 142.575, 148.925, 140.675, 157.65, 155.475, 143.35, 153.230769230769, 155.575), Y5 = c(70.775, 71.225, 70.825, 73.1499999999999, 71.925, 71.25, 73.625, 70.8, 70.1, 72.225, 70.65, 71.7999999999999, 72.25, 71.5, 70.3, 71.5, 72.5, 69.65, 69.1750000000001, 72.75), Y6 = c(615.6, 575.5, 615.4, 589.9, 606.4, 636.6, 551.55, 579.5, 589.4, 598.7, 470.15, 488.8, 512.5, 474.1, 500.75, 524.7, 504.9, 484.1, 497.25, 489), Y7 = c(335.539195565588, 359.834917608241, 335.575951966647, 357.790692959951, 333.286528104675, 353.588506032236, 364.366872182404, 366.16286093936, 351.99769389574, 352.409644879114, 302.158050528416, 314.464169350194, 278.31031721728, 313.875605554085, 279.951178433919, 300.141203224282, 308.802753249051, 292.681876126123, 308.529108917156, 317.955004237038)), row.names = c(NA, -20L), class = "data.frame")
Now, I conduct Principle component analysis.
pca<- prcomp(df[, c(-1,-2)], scale = TRUE)
fviz_pca_biplot(pca, label = "var", col.var="Black",labelsize = 4,
arrowsize = 0.3, pointsize=2, repel = TRUE, habillage = df$SEASON,
addEllipses = TRUE, palette = c("grey25", "grey65"))
labs(title="")
scale_x_continuous(breaks= seq(-4,4,1), limits = c(-4,4))
theme_grey(base_size=16, base_family="serif")
theme(axis.line= element_line(size=0.5, colour="black"),
axis.title.y= element_text (margin = margin(t=0, r=0, b=0, l=0)),
legend.position = c(0.9,0.1),
legend.title = element_blank(),
legend.key = element_rect(color = "white", fill = "white"),
legend.background= element_rect(fill= alpha("grey",.05)),
plot.margin= margin(-0.5,0.5,0,0.3,"cm"))
windows(width=6, height=5.5)
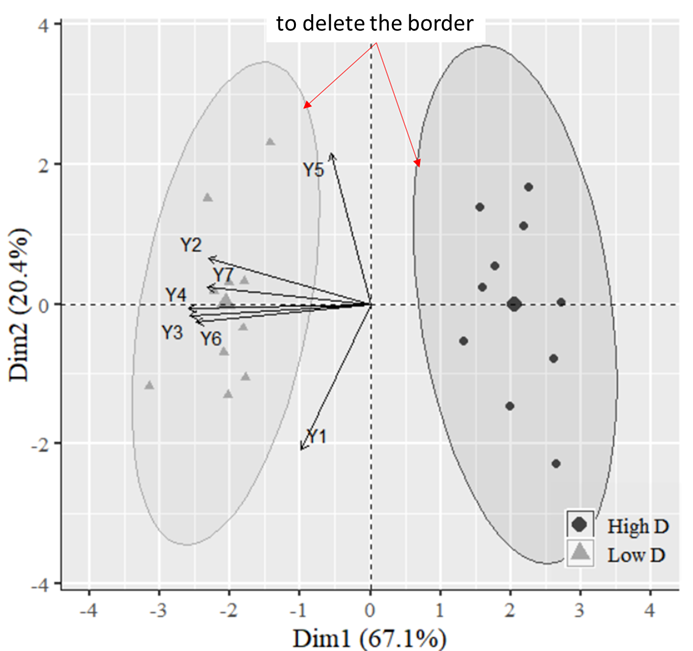
This is a graph for PCA.
Now, I'd like to
- delete the border of circles.
- provide transparent color in circles (i.e. 60% transparent in blue at Low D).
- change the point shape to round in both groups (At Low D, it's triangle now).
Could you tell me how to do it?
Many thanks!!
CodePudding user response:
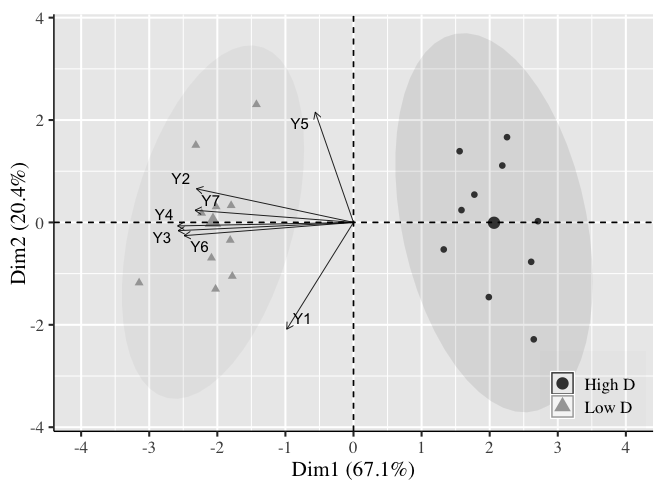
To change the borderline of the ellipses, you could use ggplot_build and change the linetype to "blank" like this:
library(factoextra)
library(ggplot2)
p <- fviz_pca_biplot(pca, label = "var", col.var="Black",labelsize = 4,
arrowsize = 0.3, pointsize=2, repel = TRUE, habillage = df$SEASON,
addEllipses = TRUE, palette = c("grey25", "grey65"))
labs(title="")
scale_x_continuous(breaks= seq(-4,4,1), limits = c(-4,4))
theme_grey(base_size=16, base_family="serif")
theme(axis.line= element_line(size=0.5, colour="black"),
axis.title.y= element_text (margin = margin(t=0, r=0, b=0, l=0)),
legend.position = c(0.9,0.1),
legend.title = element_blank(),
legend.key = element_rect(color = "white", fill = "white"),
legend.background= element_rect(fill= alpha("grey",.05)),
plot.margin= margin(-0.5,0.5,0,0.3,"cm"))
q <- ggplot_build(p)
q$data[[2]]$linetype = "blank"
q <- ggplot_gtable(q)
plot(q)
Created on 2022-09-10 with reprex v2.0.2