I have adata.frame such as:
df <- data.frame(region = c(rep("r1", 5), rep("r2", 5)),
family = c("A1", "A1", "A2", "A2", "A3",
"B1", "B6", "B11", "B2", "B2"),
genus = c("a1", "a2", "a1", "a1", "a1",
"b1", "b3", "b3", "b1", "b4"),
spp = c("a1a", "a4a", "a2a", "a1a", "a1a",
"b1b", "b6b", "b6b", "b2b", "b3b"))
I'm trying to create a grouped bar chart such as the following image
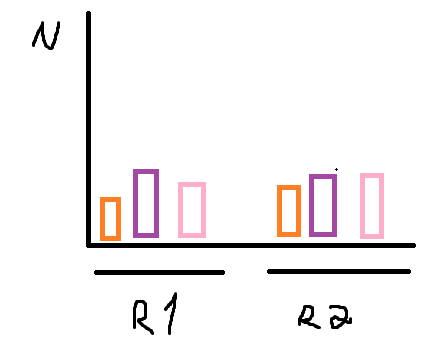
Where each color is respectively family, genus, and spp and "R1"/"R2 are region. The N axis must be the n_distinct() of family, genus, and spp. How can I approach this?
CodePudding user response:
Get your data into shape:
library(ggplot2)
library(tidyr)
plot_df <- df |>
group_by(region) |>
summarise(across(family:spp, n_distinct)) |>
pivot_longer(-region)
plot_df
# A tibble: 6 x 3
# region name value
# <chr> <chr> <int>
# 1 r1 family 3
# 2 r1 genus 2
# 3 r1 spp 3
# 4 r2 family 4
# 5 r2 genus 3
# 6 r2 spp 4
Then you can plot it:
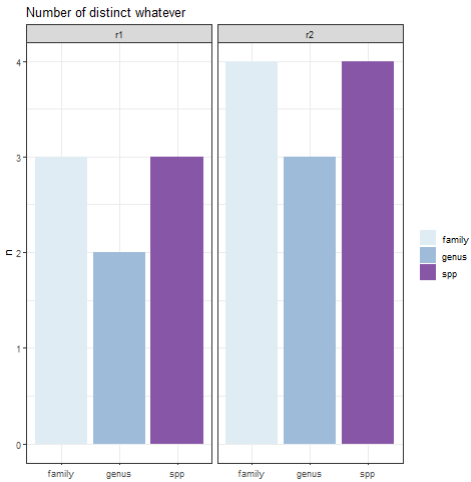
ggplot(plot_df)
geom_col(mapping = aes(
x = name,
y = value,
fill = name
))
facet_wrap(vars(region))
labs(
y = "n",
title = "Number of distinct whatever"
)
theme_bw()
theme(
axis.title.x = element_blank(),
legend.title = element_blank()
)
scale_fill_brewer(palette=3)
Edit: made the plot look slightly nicer.
CodePudding user response:
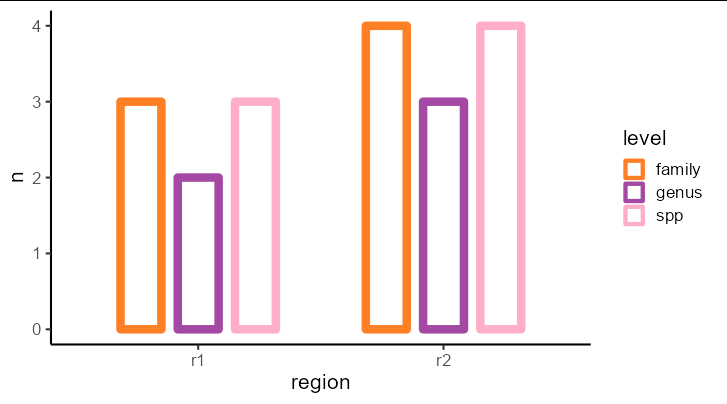
You can plot region directly on the x axis without faceting too:
library(tidyverse)
pivot_longer(df, family:spp, names_to = 'level') %>%
group_by(region, level) %>%
summarize(n = n_distinct(value)) %>%
ggplot(aes(region, n, color = level))
geom_col(position = position_dodge(width = 0.7), width = 0.5,
fill = 'white', size = 3)
theme_classic(base_size = 16)
scale_color_manual(values = c('#ff7f27', '#a349a4', '#ffaec9'))